Polish Scientists Contribute to Breakthrough Study on tRNA Modifications Published in EMBO Journal
Researchers from Kraków and Warsaw, in partnership with collaborators from UK and France, have demonstrated how the pseudouridine modification enhances the structural integrity of human tRNAs. These insights may contribute to developing new RNA-based therapies and broaden our overall understanding of RNA biology.
In a recent study, scientists from the Małopolska Centre of Biotechnology (Jagiellonian University) in Kraków, in collaboration with researchers from the International Institute of Molecular and Cell Biology (IIMCB) in Warsaw and institutions from the United Kingdom and France, focused on transfer RNAs (tRNAs) – essential molecules that decode genetic information into proteins.
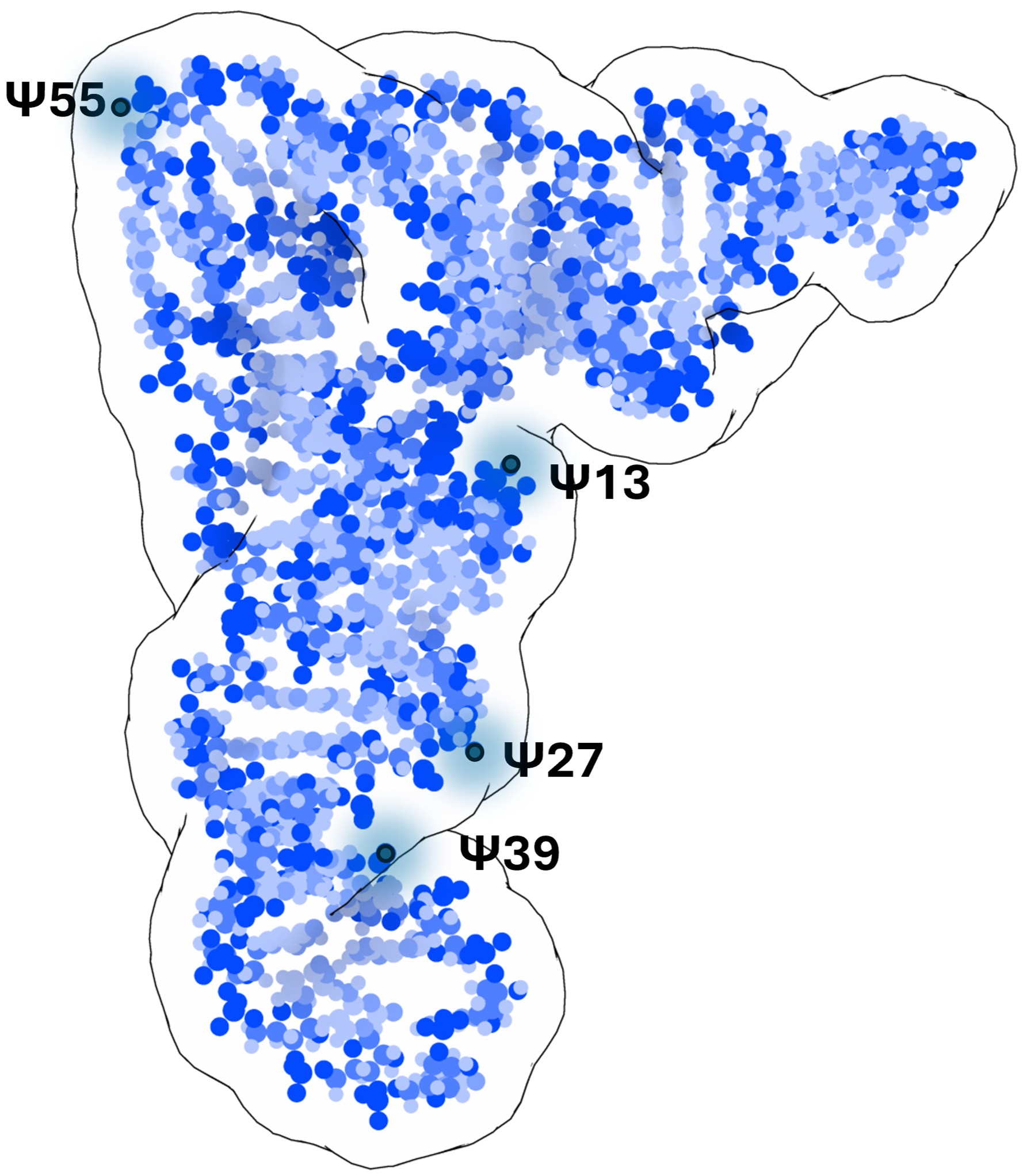 tRNA structure modeled into a cryo-EM map, with positions of uridine to
tRNA structure modeled into a cryo-EM map, with positions of uridine to
pseudouridine modifications indicated
Decoding the Structural Impact of RNA Modifications
Structural studies of tRNAs have long posed a challenge due to their small size and complex architecture. This research analyzed four human tRNAs before and after the enzyme-mediated introduction of pseudouridine (Ψ), revealing that this specific modification enhances molecular stability and induces local structural changes. In particular, the study identified interactions between the D- and T-arms of tRNAs as critical for maintaining their overall tertiary structure.
“We demonstrated the profound impact of RNA modifications on tRNA stability and function,” said Prof. Sebastian Glatt, the leader of the project. “Our findings not only improve our understanding of tRNA biogenesis but also highlight the potential applications of engineered tRNAs in therapeutic contexts.”
KEY RESULTS FROM THE STUDY:
• Cryo-EM structural determination of multiple unmodified and pseudouridylated human tRNAs
• Clear evidence that specific pseudouridine modifications, especially at positions 13 and 55, significantly increase the thermal stability of tRNAs.
• Context-dependent impacts of modifications, indicating not all pseudouridine modifications are equally beneficial.
Interdisciplinary Approach: From Cryo-EM to Computational Modeling
The IIMCB team, led by Prof. Janusz Bujnicki, contributed essential computational modeling and molecular dynamics simulations that provided pivotal insights into the mechanisms underlying tRNA stabilization. “This study illustrates the power of computational modeling for interpreting low- to medium-resolution cryo-EM maps,” said Prof. Bujnicki, co-leader of the study. “Our simulations clearly show how pseudouridine stabilizes key interactions within tRNA structures, paving the way for future analyses of other RNA modifications.”
The research was made possible through a unique interdisciplinary approach combining experimental and computational techniques. It employed advanced single-particle cryo-electron microscopy (cryo-EM), biophysical assays, and molecular modeling.
 Members of Prof. Bujnicki’s group at IIMCB (Laboratory of Bioinformatics
Members of Prof. Bujnicki’s group at IIMCB (Laboratory of Bioinformatics
and Protein Engineering) who participated in the study. From left: S.
Naeim Moafinejad, Dr. Sunandan Mukherjee, and Dr. Satyabrata Maiti
Funding for this study was provided by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program, the National Science Center and Poland's PLGrid high-performance computing infrastructure.
The article, titled “Determining the effects of pseudouridine incorporation on human tRNAs”, is freely available at: https://www.embopress.org/doi/full/10.1038/s44318-025-00443-y.

