Scientists from the Laboratory of Protein Structure and Institute of Biophysics of the Czech Academy of Sciences published an article in Nature Communications which describes the mechanism of RuvC Holiday junction resolvase. The article describes how RuvC resolvase uses dynamic probing of the Holliday junction to achieve sequence specificity and efficient resolution. The bacterial protein RuvC is a canonical resolvase that introduces two symmetric cuts into the HJ. For complete resolution of the HJ, the two cuts need to be tightly coordinated. They are also specific for cognate DNA sequences. Using a combination of structural biology, biochemistry, and a computational approach, Karolina Górecka, Aleksandra Szlachcic and Marcin Nowotny from IIMCB with Miroslav Krepl and Jiri Sponer from Institute of Biophysics of the Czech Academy of Sciences show that correct positioning of the substrate for cleavage requires conformational changes within the bound DNA. These changes involve rare high-energy states with protein-assisted base flipping that are readily accessible for the cognate DNA sequence but not for non-cognate sequences. These conformational changes and the relief of protein-induced structural tension of the DNA facilitate coordination between the two cuts. The unique DNA cleavage mechanism of RuvC shows the importance of high-energy conformational states in nucleic acid readouts. This highly interdisciplinary work also demonstrated the power of combining experimental and computational know-how of the two collaborating groups.
Górecka KM&, Krepl M*,&, Szlachcic A, Poznański J, Šponer J, Nowotny M*. RuvC uses dynamic probing of the Holliday junction to achieve sequence specificity and efficient resolution. Nat Commun. 2019 Sep 10;10(1):4102. *corresponding authors, &co-first authors
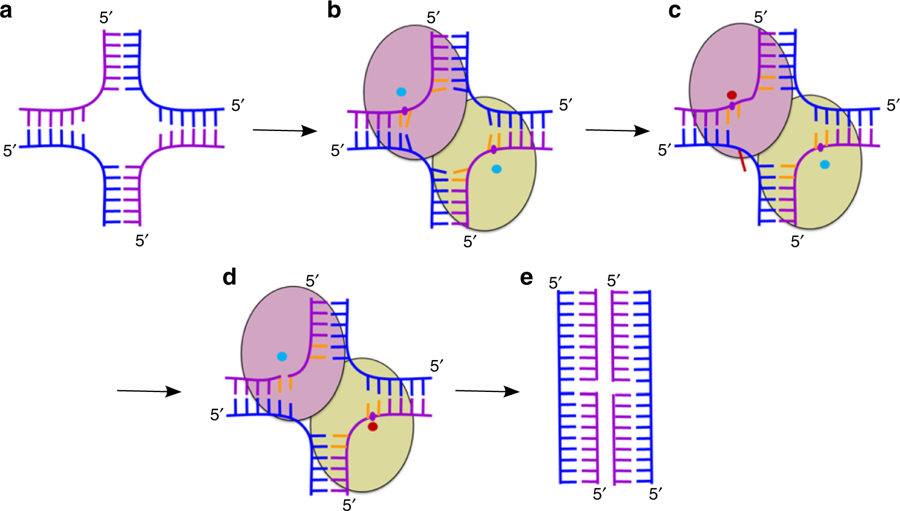
Cartoon representation of the mechanism of HJ resolution by RuvC. a Holliday junction. Cleaved and non-cleaved DNA strands are shown in purple and blue ladder-like representations, respectively. b Binding of the HJ DNA. The subunits of the dimer are shown as yellow green and pink ovals. The scissile phosphate is marked as a purple circle. Cyan circles show active sites in an inactive configuration. c Flipping of the adenine (red) opposite the scissile base. The active site in the catalytic configuration is shown as a red circle. d The second cut. e Resolution products
