Scientists from the Laboratory of Protein Structure published an article in which they describe a structural and mechanistic characterization of the mitochondrial exoribonuclease complex (mtEXO) which is the main executor of RNA degradation in yeast mitochondria. It is composed of Dss1 3ʹ-to-5ʹ exoribonuclease and Suv3 helicase that act in concert and efficiently remove defective RNAs and excised introns. Crystal structure of Dss1 from Candida glabrata revealed that it is a unique member of the RNase II family with specialized domains responsible for interactions with Suv3 helicase. The arrangement of both subunits revealed by the crystal structure of the complex enables the helicase motor to feed the 3ʹ end of the RNA into the catalytic channel of Dss1 for effective degradation. This very tight co-operation of helicase and nuclease activities within the complex is unprecedented and is particularly important for degradation of structured RNAs which cannot be processed by Dss1 on its own and for which the unwinding activity of Suv3 is required.
Razew M, Warkocki Z, Taube M, Kolondra A, Czarnocki-Cieciura M, Nowak E, Labedzka-Dmoch K, Kawinska A, Piatkowski J, Golik P, Kozak M, Dziembowski A, Nowotny M. Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components. Nat Commun. 2018 Jan 8;9(1):97.
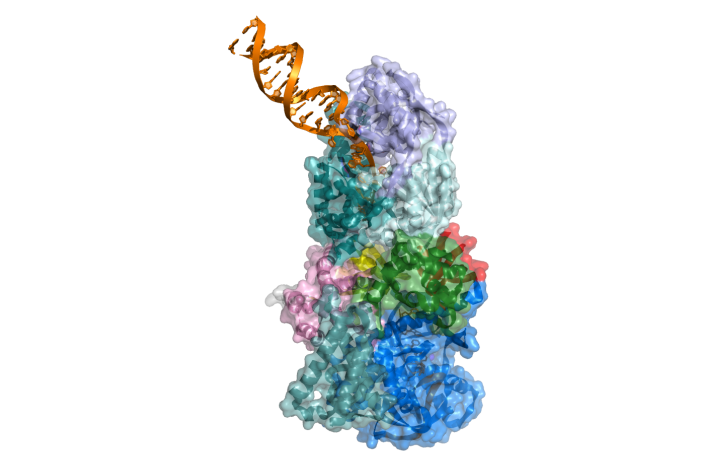
Figure: A model of the Candida glabrata mtEXO complex in action. The Suv3 helicase (top) unwinds the dsRNA (orange) and feeds a free 3′ end to the catalytic site of the exoribonuclease Dss1 (bottom) where the substrate is degraded.
