We already know the results of the Best Papers Awards 2022! The best scientific papers of the past year, affiliated with IIMCB, were awarded. Any experimental work of a scientific nature with no subject restriction could be submitted to this Competition. The best publications were selected by the Jury consisting of all Laboratory Leaders, based on content and significance and not bibliometric data. Laboratory Leaders were not allowed to vote for papers from their own laboratory. In 2022, the following papers were awarded:
- 1st PLACE
Kaczmarska Z*, Czarnocki-Cieciura M*, Górecka-Minakowska KM*, Wingo RJ, Jackiewicz J, Zajko W, Poznański JT, Rawski M, Grant T, Peters JE#, Nowotny M#. Structural basis of transposon end recognition explains central features of Tn7 transposition systems. Mol Cell, 2022; 82(14):2618-32.e7. doi: 10.1016/j.molcel.2022.05.005
*these authors contributed equally; #corresponding authors; in bold authors affiliated with IIMCB
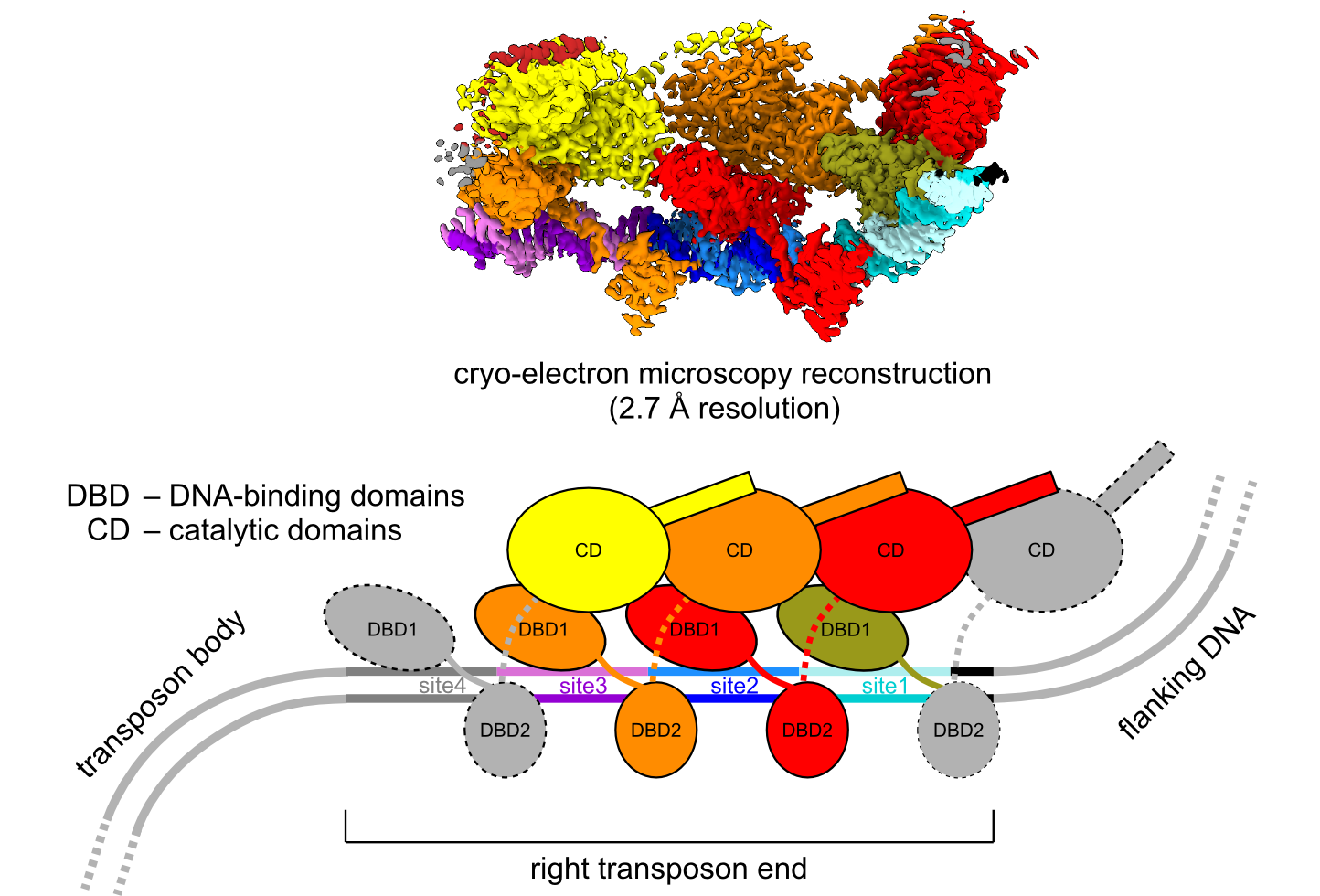
Transposons, also called “jumping genes”, are DNA fragments that can move within or between genomes in a process called transposition. In bacteria, transposons are involved in the transmission of antibiotic resistance and virulence genes. Bacterial Tn7 elements are among the best-studied and most widespread DNA transposons. Tn7 mobility is mediated by five element-encoded proteins. Transposition occurs via a cut-and-paste mechanism that is executed by a heteromeric transposase, TnsA-TnsB, which is recruited to the target DNA by TnsC protein, which is an AAA+ ATPase. TnsC interacts with one of the two target selectors, TnsD or TnsE. TnsD directs the element to the conserved chromosomal attTn7 site, whereas TnsE allows transposition to conjugal plasmids. CRISPR-associated transposon (CAST) elements that use element-encoded CRISPR-Cas systems for RNA-guided DNA transposition are related to Tn7 and encode TnsB-like transposases. They may provide new tools for next-generation gene editing.
Scientists from the Laboratory of Protein Structure, led by Marcin Nowotny, in collaboration with the Joe Peters group from Cornell University, studied the structure and mechanism of prototypic E. coli Tn7 TnsB. They used cryoelectron microscopy (cryo-EM) to determine the structure of a complex of TnsB with double-stranded DNA that corresponded to the right end of the transposon at 2.7 Å resolution. The structure shows that multiple TnsB chains, which adopt a beads-on-a-string architecture, interact with repeating binding sites in the DNA. Upon this interaction the DNA-binding and catalytic domains of TnsB chains are arranged in a tiled and intertwined fashion. TnsB forms few base-specific contacts with DNA that lead to binding preference rather than strict specificity. The formation of an array of TnsB molecules that bind to multiple weakly conserved sites at appropriate spacing converts this preference into specific end recognition. These scientists also proposed a model of the TnsB strand-transfer complex that aims to understand late steps of the Tn7 TnsB reaction. Collectively, these results help explain how subtle differences in the spacing of binding sites are used for specific transposon end recognition and define central features of Tn7 transposition systems.
- 2nd PLACE
Das A, Thapa P, Santiago U, Shanmugam N, Banasiak K, Dąbrowska K, Nolte H, Szulc NA, Gathungu RM, Cysewski D, Krüger M, Dadlez M, Nowotny M, Camacho CJ, Hoppe T, Pokrzywa W#. A heterotypic assembly mechanism regulates CHIP E3 ligase activity. EMBO J, 2022; 41(15):e109566, doi: 10.15252/embj.2021109566
#corresponding author; in bold authors affiliated with IIMCB
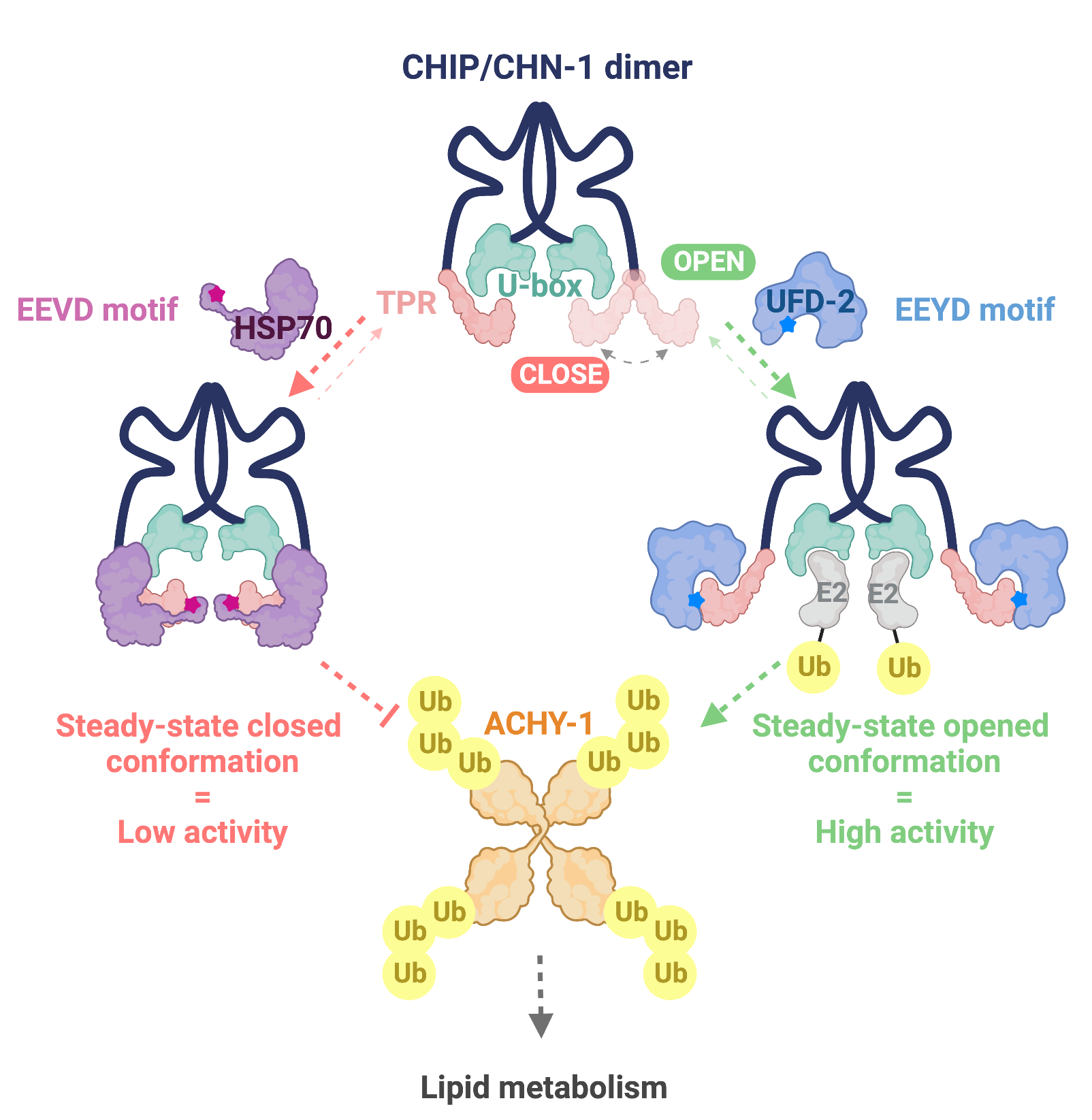
The fate of eukaryotic proteins is supervised by the chaperone network and the ubiquitin-proteasome system (UPS). CHIP (C‐terminus of Hsc70‐interacting protein) is an important quality control E3 ubiquitin ligase that links the chaperone system with the UPS to degrade damaged proteins. It also mediates chaperone-independent ubiquitylation and can interact with other E3s. However, the regulation of CHIP processivity and substrate selectivity in response to chaperone and E3 binding has remained unclear. Scientists from the Laboratory of Protein Metabolism, led by Wojciech Pokrzywa, performed a structural-functional analysis of the complex that was formed by CHIP and UFD-2, another E3 ubiquitin ligase, guided by the idea that they form a highly processive ubiquitylation system alternative to the CHIP/chaperone axis. The data showed that UFD-2 binding promotes structural gain of function in CHIP. The researchers demonstrated that the heat shock protein Hsp70 outcompetes UFD-2 for CHIP binding and negatively regulates activity of the complex by stabilizing the auto-inhibited state of CHIP. Using the nematode Caenorhabditis elegans, the scientists discovered that an interaction with UFD-2 enables CHIP to regulate S-adenosylhomocysteinase, an enzyme that is crucial for cellular methylation. The results obtained by the Pokrzywa group open new horizons in research on the cooperation of ubiquitin ligases in gaining high activity and substrate selectivity. In addition, the revealed CHIP processivity switching mechanism has potential application in targeted protein degradation approaches.
- 3rd PLACE
Niescierowicz K*, Pryszcz L*, Navarrete C*, Tralle E*, Sulej A*, Abu Nahia K, Kasprzyk ME, Misztal K, Pateria A, Pakuła A, Bochtler M#, Winata C#. Adar-mediated A-to-I editing is required for embryonic patterning and innate immune response regulation in zebrafish. Nat Commun, 2022; 13(1):5520, doi: 10.1038/s41467-022-33260-6
*these authors contributed equally; #corresponding authors; in bold authors affiliated with IIMCB
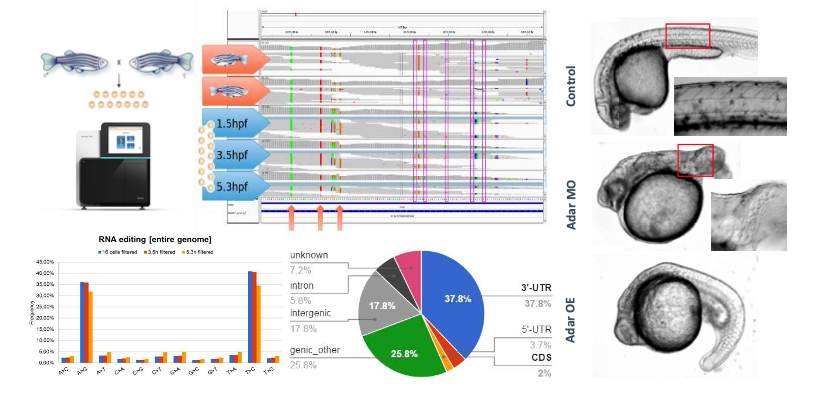
Adenosine-to-inosine (A-to-I) editing is necessary for regulating the innate immune system in humans and other mammals and it is implicated in human diseases, including autoimmune conditions. The enzyme adenosine deaminase acting on RNA (Adar) is responsible for catalyzing such editing, which entails the deamination of adenosine (A) at the C6 position, giving rise to an inosine (I). Researchers from the Laboratory of Zebrafish Developmental Genomics and Laboratory of Structural Biology, led by Cecilia Winata and Matthias Bochtler, respectively, investigated the role of Adar in zebrafish, where it is highly expressed in the earliest stages of embryogenesis. Genome-wide editing discovery by combined analyses of the parental genome and embryonic transcriptome uncovered prevalent A-to-I editing in maternal and the earliest zygotic transcripts, the majority of which occurred in the 3’-untranslated region. Transcripts that are known to play a role in gastrulation and embryonic patterning were found to contain multiple editing sites, suggesting that Adar may exert its function through them. Through Adar loss- and gain-of-function experiments, the researchers demonstrated that maternal Adar function is essential for proper embryonic patterning along the antero-posterior and dorso-ventral axes, and this function depends on an intact deaminase domain. Analyses of adar zygotic mutants revealed the distinct zygotic function of Adar in regulating the innate immune response, a role that is conserved in mammals. Collectively, the study established a novel function of Adar-mediated A-to-I editing in regulating embryonic patterning and revealed the conservation of zygotic Adar function between zebrafish and mammals.