Publications in Nature Communications and Nucleic Acids Research. We are gaining a better understanding of life at the cellular level.
Researchers from the Laboratory of Protein Metabolism at the International Institute of Molecular and Cell Biology in Warsaw (IIMCB) have published two articles in prestigious scientific journals. The discoveries featured in the Editors' Highlights section of Nature Communications open new possibilities for research on social interactions and their impact on biological systems at the cellular level. The second publication in Nucleic Acids Research discusses the unique web server DEGRONOPEDIA, which is a valuable tool for researchers in biochemistry, molecular biology, and medicine.
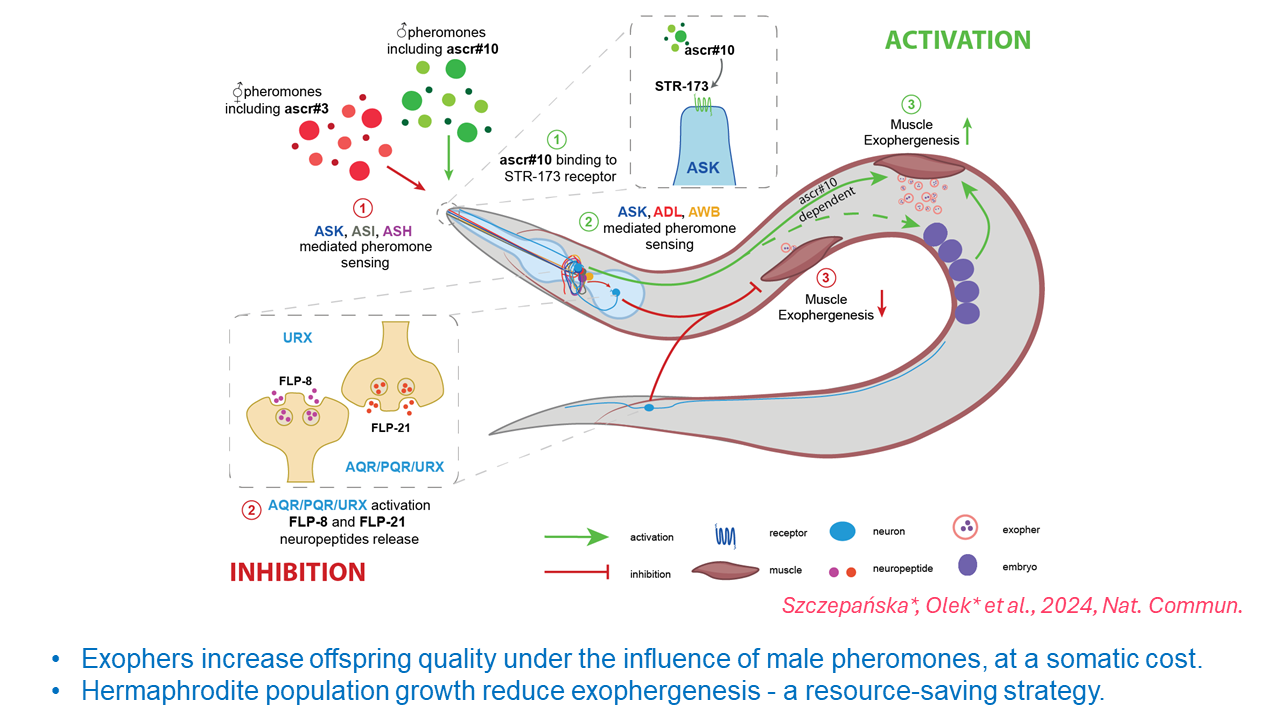
Advances in molecular biology are increasingly clarifying the mechanisms controlling life processes at the molecular level. One such achievement is the study concerning exophers, or extracellular vesicles of muscles in the model organism C. elegans, which plays a crucial role in biomedical research. The results of these studies, published in Nature Communications by Dr. Wojciech Pokrzywa's team from the IIMCB and Dr. Michał Turek from the Institute of Biochemistry and Biophysics, Polish Academy of Sciences, focus on the role that olfactory neurons and gender-specific pheromones play in regulating exopher production. These diverse chemical compounds, also secreted by nematodes, are crucial for intercellular communication.
The researchers discovered that communication through specific pheromones influences the production of exophers by muscles. Under the influence of male pheromones, these muscles produce an increased amount of vesicles, which supports the development of embryos in the mother's uterus, but this is associated with a somatic cost to the mother's body. Additionally, population growth, also sensed via pheromones, may result in reduced exopher production, which constitutes a resource-saving strategy.
G protein-coupled receptors, lesser-known classes of neurons, and their neuropeptides play a crucial role in this process, revealing a complex network of neural communication responsible for regulating exopher production in response to social signals.
"This discovery opens new perspectives for understanding how environmental signals can affect the functioning of organisms at the cellular level, including humans," emphasizes Dr. Wojciech Pokrzywa from IIMCB.
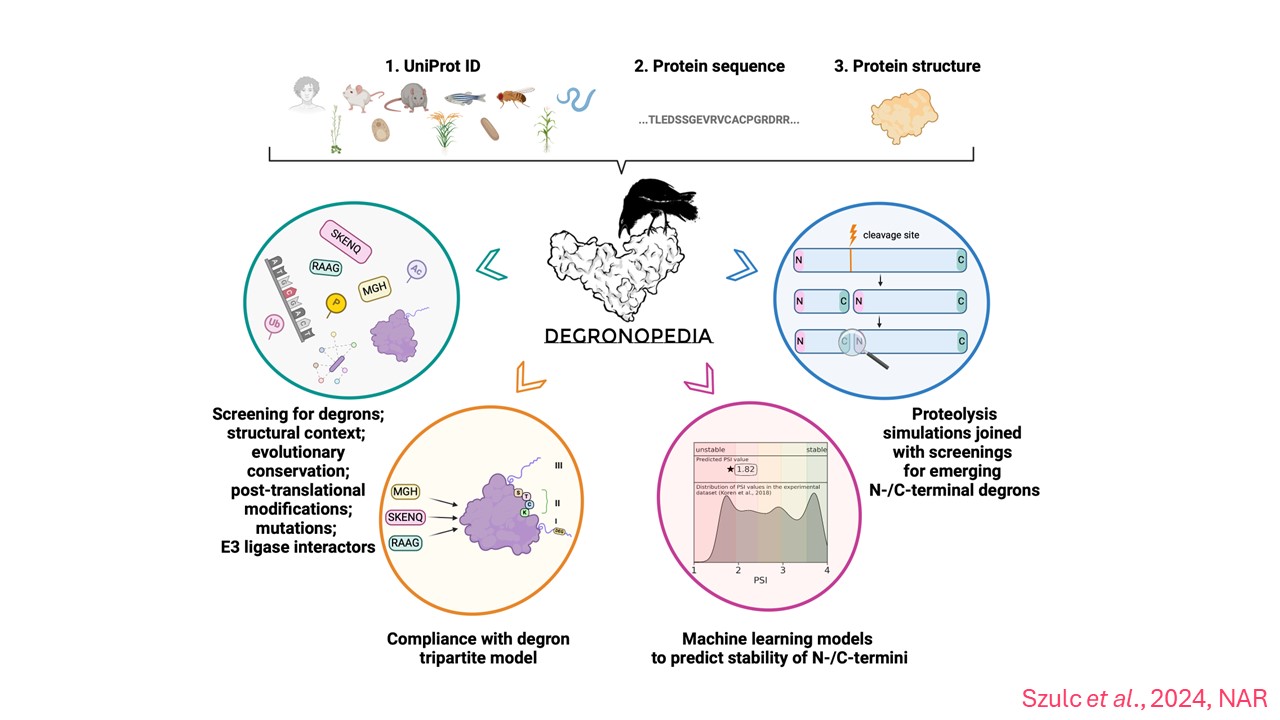
The second publication, also authored by Dr. Wojciech Pokrzywa's team and published in Nucleic Acids Research, is devoted to DEGRONOPEDIA. It is an advanced online platform that allows the analysis of degrons - short, linear protein motifs recognized by E3 ubiquitin ligases. Degrons are crucial in the protein signaling process, leading to their ubiquitination and degradation, essential for "cleaning" the cell from unnecessary or defective proteins, ensuring its proper function.
Despite intensive research on the functioning of degrons, there has been a lack of resources enabling their systematic analysis until now. DEGRONOPEDIA was created to fill this gap, offering the ability to search the proteome - the complete catalog of proteins in a given organism - for degrons and map them in the context of ubiquitination. The server also provides information about protein sites that may undergo ubiquitination and their disordered regions, which may play a role during degradation. Additionally, the platform provides data on the evolutionary conservation of degrons, their modifications, and mutations.
"It is worth noting that DEGRONOPEDIA uses machine learning technologies to identify degrons, e.g. at newly formed protein ends during proteolysis, that is protein cleavage. The practical application of the platform has been confirmed by experimental studies," says the article's co-author and project leader, Natalia Szulc, a PhD student from the Laboratory of Protein Metabolism at IIMCB.
DEGRONOPEDIA is available for free at https://degronopedia.com/
The article "Pheromone-based communication influences the production of somatic extracellular vesicles in C. elegans" is available at: https://www.nature.com/articles/s41467-024-47016-x
You can read the article "DEGRONOPEDIA: a web server for proteome-wide inspection of degrons" at: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae238/7639542