Scientists investigated how bacteria defend themselves against viruses
Researchers from the International Institute of Molecular and Cell Biology in Warsaw together with scientists from Department of Experimental Medical Science, Lund University, have described a detailed study of the AbiA protein, an enzyme involved in bacterial defense against viral infections. The structural and biochemical analyses of the proteins, published in Nucleic Acid Research and supplemented by in vivo studies, shed light on the previously unexplored mechanism of AbiA's action.
Bacteria, like all living organisms, are susceptible to viral infections. To prevent them or to stop viral replication after infection, microorganisms employ various defense strategies. Many bacterial proteins involved in antiviral defense are classified as reverse transcriptases (RT). Typical RTs use RNA as a template to create copies of genetic information in the form of DNA. Certain bacterial RTs are believed to be responsible for programmed cell death, known as abortive infection. Thus, an infected bacterium "commits suicide" to stop the replication of the virus.
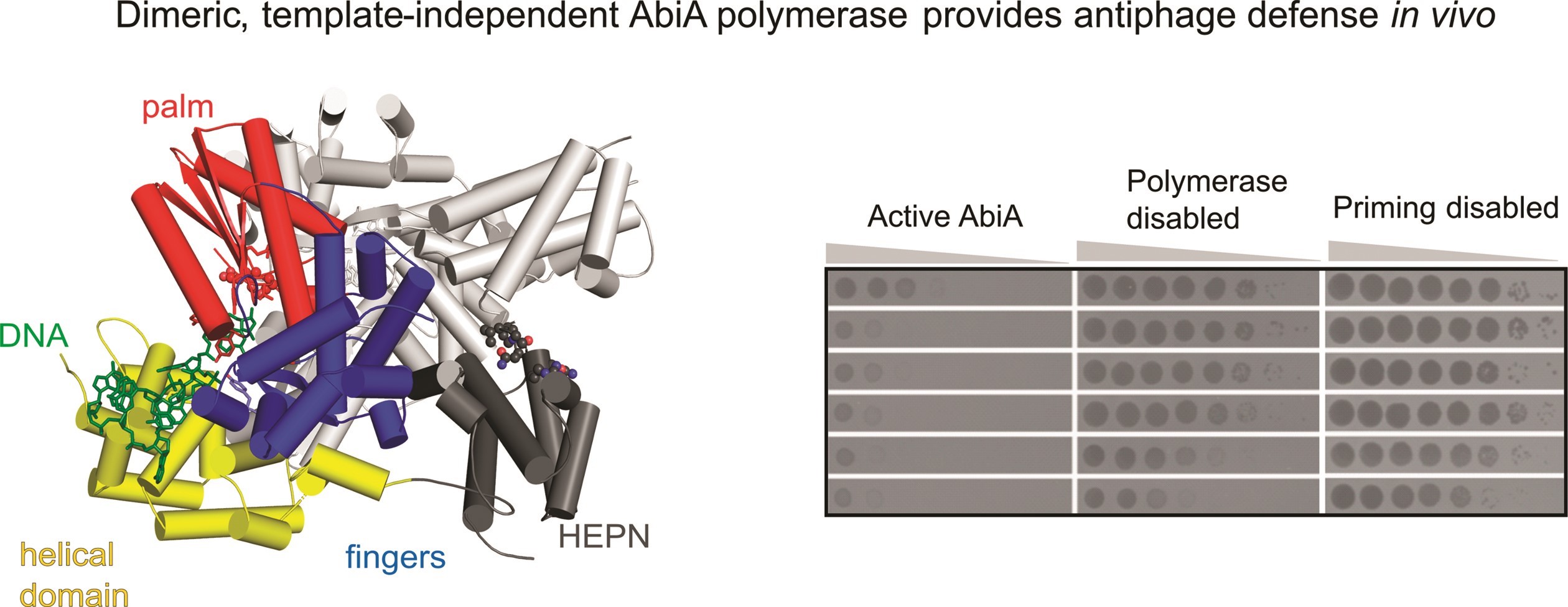
In the study, the team from the Laboratory of Protein Structure at IIMCB, led by Marcin Nowotny, determined the first atomic three dimensional structures of one of the bacterial RTs, the AbiA protein, bound to DNA produced by this enzyme. The research revealed significant differences between AbiA and other Abi-type transcriptases. AbiA forms a dimer (a combination of two identical molecules). Besides the RT-like domain, which is responsible for DNA production, and an α-helical domain, which stabilizes the produced DNA, AbiA also contains a HEPN domain common to many antiviral proteins. In vitro experiments studying the enzyme's activity showed that AbiA initiates DNA synthesis using a protein-priming mechanism (the first nucleotide of the produced DNA strand is attached to one of the protein's residues, rather than to an existing fragment of nucleic acid as in most cases) and can produce DNA fragments of about 100 nucleotides in length.
Sequence analysis of these DNA products, conducted by researchers from the Laboratory o RNA Biology, revealed that the enzyme has a strong preference for adenosine and cytidine (two of the four basic nucleotides in DNA), in contrast to the AbiK enzyme, which produces much longer products with completely random sequences. Additionally, viral infection tests conducted by collaborating researchers from Lund University showed that DNA polymerase activity is required for AbiA's antiphage action. These results also suggest that, contrary to previous assumptions, the defense mechanism of AbiA does not involve triggering the suicide of the infected cell.
The article "Structure-functional characterization of Lactococcus AbiA phage defense system" can be accessed at this link: https://academic.oup.com/nar/article/52/8/4723/7642066?searchresult=1
Caption: General structure of the L. lactis AbiA-DNA complex (left) and antiphage activity test of wild-type AbiA and its mutated variants (right).