Mathematical modelling sheds light on the origins of an aggressive leukemia subtype
Researchers from the International Institute of Molecular and Cell Biology in Warsaw (IIMCB), together with partners in Heidelberg, Kiel, and Dresden, have investigated the sequence of genetic events that drive one of the most aggressive forms of acute myeloid leukemia (AML). Their study, published in Leukemia (Springer Nature), explains the damage to the TP53 gene and the development and interactions of large-scale chromosomal abnormalities in this rare leukemia subtype.
The work focused on a severe and insufficiently understood form of leukemia known as complex karyotype AML (CK-AML). Although rare, CK-AML is a very serious form of the disease – many patients do not survive beyond one year, and no effective treatment is currently available. In this research project, the scientists combined genetic data with mathematical modelling to uncover molecular mechanisms of TP53 gene defects that shape the disease and contribute to its clinical severity.
TP53: The guardian protein at the center of the disease
TP53 is a gene that encodes the p53 protein, often referred to as the “guardian of the genome”. It halts the division of damaged cells and triggers DNA repair or apoptosis – a programmed form of cell death that eliminates cells with harmful mutations. When TP53 is damaged, this control is weakened, allowing abnormal cells to divide unchecked, increasing the risk of cancer development.
TP53 defects arise in many cancer types, including breast, lung, and colorectal cancers, as well as leukemias. Loss of proper p53 function promotes chromosomal instability, facilitating the emergence of complex karyotypes that characterize the most aggressive forms of AML. For this reason, TP53-mutated CK-AML is considered a distinct biological subtype of the disease, following its own accelerated evolutionary trajectory.
Scientific curiosity driven by patients’ needs
For the study’s first author, Anna Fedenko from IIMCB’s Laboratory of Structural Biology, the project reflects her scientific curiosity about molecular mechanisms in complex cancers. When Prof. Matthias Bochtler, Head of the Laboratory of Structural Biology and corresponding author of the study, proposed a sequencing-based investigation into TP53-mutated CK-AML, her decision to join was immediate.
“I knew I wanted to take part in this project and see it through, as my heart has always been leaning toward precision medicine and its immense potential to translate scientific findings into direct benefits for patients,” says Anna Fedenko.
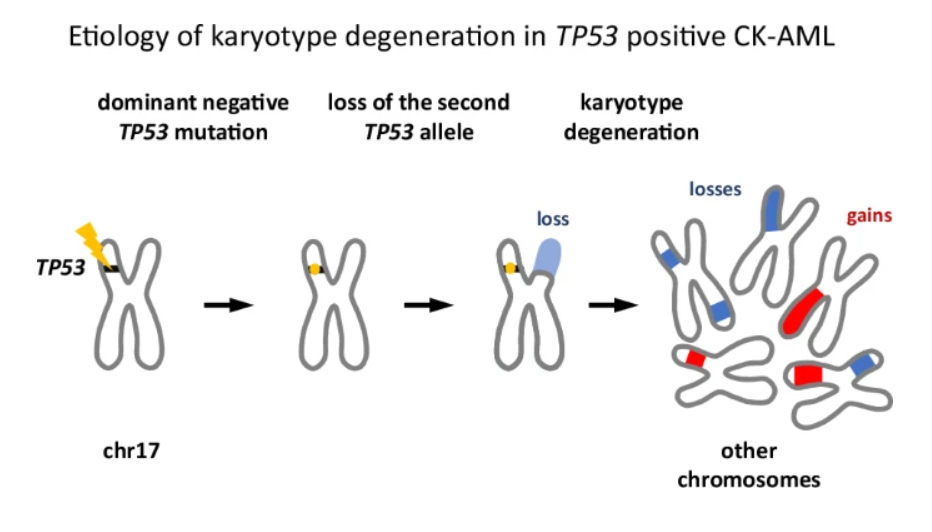 Fig.: First, one copy of the TP53 gene becomes mutated, which weakens its ability to protect the DNA. Then, the mutation promotes the deletion of the second, still-healthy TP53 copy. At this point, the cell has no working TP53 left. Without TP53, the genome becomes unstable. As a result, many additional DNA changes appear, such as missing pieces of chromosomes, extra copies, or chromosomes breaking and re-joining incorrectly. These many changes together create genomic chaos, which is what we call a complex karyotype.
Fig.: First, one copy of the TP53 gene becomes mutated, which weakens its ability to protect the DNA. Then, the mutation promotes the deletion of the second, still-healthy TP53 copy. At this point, the cell has no working TP53 left. Without TP53, the genome becomes unstable. As a result, many additional DNA changes appear, such as missing pieces of chromosomes, extra copies, or chromosomes breaking and re-joining incorrectly. These many changes together create genomic chaos, which is what we call a complex karyotype.
A combined sequencing-and-modelling strategy
Working on a rare disease meant facing the limitations of small patient cohorts, yet the analytical strategy allowed the team to extract far more information than expected. Bone marrow samples from the patients were analyzed using next-generation sequencing (NGS) in collaboration with Genomics Core Facility at Centre of New Technologies (CeNT) and , enabling the simultaneous detection of point mutations and chromosomal-level abnormalities in a single run.
The data were integrated with a simple mathematical model that estimated the proportions of cells carrying specific TP53 mutations and copy-number alterations. By comparing variant allele frequencies (VAF) with copy-number variation (CNV), the researchers inferred how different genetic events accumulated over time – a process not directly observable in patients.
TP53-Mutated AML emerges as a distinct disease entity
A key moment in the research came as the analytical layers converged. “There is a point when the results align and the broader picture becomes clear”, Anna Fedenko reflects. “Seeing how the findings fit together — and how many new questions they raise — was one of the most rewarding stages of this work”.
The study aligns with the opinion that TP53-mutated acute myeloid leukemia with a complex karyotype is a distinct disease subtype, with unique pathophysiology and a specific order of genetic events. This can improve diagnostic classification and treatment choices in AML and may enhance prognosis and risk assessment. Furthermore, the scientific teams’ in-depth examination of the sequence of events leading to total TP53 inactivation can aid in developing new therapies that aim at TP53-restoring mechanisms.
BROADER IMPLICATIONS OF THE RESEARCH:
-
- Hematology, clinical oncology, cancer genetics – through refined understanding of the prognostic significance of TP53 defects and their prevalence in bone marrow and blood.
- Diagnostics – the research is a step towards the development of clinical-grade genomic tests and bioinformatic tools that reconstruct gene damage sequences in cancer evolution.
- Bioinformatics – via a modelling framework that infers the order of molecular events in leukemia development.
- Pharmaceutical research – particularly for teams developing targeted therapies for TP53 defects found in the most severe cancers.
More evidence required
The IIMCB scientists emphasize that the study does not represent a breakthrough in leukemia treatment. Larger, longitudinal studies will be needed to validate the findings on AML evolution, and clinical trials would be required to assess potential therapeutic implications.
“Perhaps if we learn how to switch TP53 back on, we may one day prevent or reverse this deadly disease – but this requires far deeper investigation,” Fedenko explains. She adds: “For me, this work is a drop in a big sea of discoveries and what is yet to be discovered. After all, there would be no sea without those little drops – the contributions of knowledge it consists of. This cumulative growth is the essence of basic research.”
The article Etiology of TP53-mutated complex karyotype acute myeloid leukemia in the Leukemia journal (Springer Nature) is available here: https://doi.org/10.1038/s41375-025-02835-9
The project was conducted in collaboration with clinicians from the German Cancer Research Center (DKFZ) in Heidelberg, University of Heidelberg, Carl Gustav Carus University in Dresden, University Hospital Schleswig-Holstein in Kiel, and Heidelberg University Hospital. The authors acknowledge the support of the IIMCB and CeNT Core Facilities – including library preparation, sequencing, and cell sorting – as well as the patients and families who generously contributed samples for research.
Funding: Foundation for Polish Science (FNP),EU European Regional Development Fund [POIR.04.04.00-00-5D81/17-00]; Polish National Agency for Academic Exchange [NAWA, PPI/APM/2018/1/00034], Polish National Science Centre [NCN, 2018/30/Q/NZ2/00669], EU NextGenerationEU under National Recovery and Resilience Plan, EU), European Union (RACE Project no 101059801) and European Funds for Smart Economy 2021-2027 (FENG) (RACE-PRIME, IRAP FNP program) and supported by IIMCB IN-MOL-CELL Infrastructure (RRID:SCR_021630).


