Scientists from the Laboratory of Biomolecular Interactions and Transport AMU/IIMCB, led by Jan Berezovsky, PhD, has recently published a paper entitled “TransportTools: a library for high-throughput analyses of internal voids in biomolecules and ligand transport through them”.
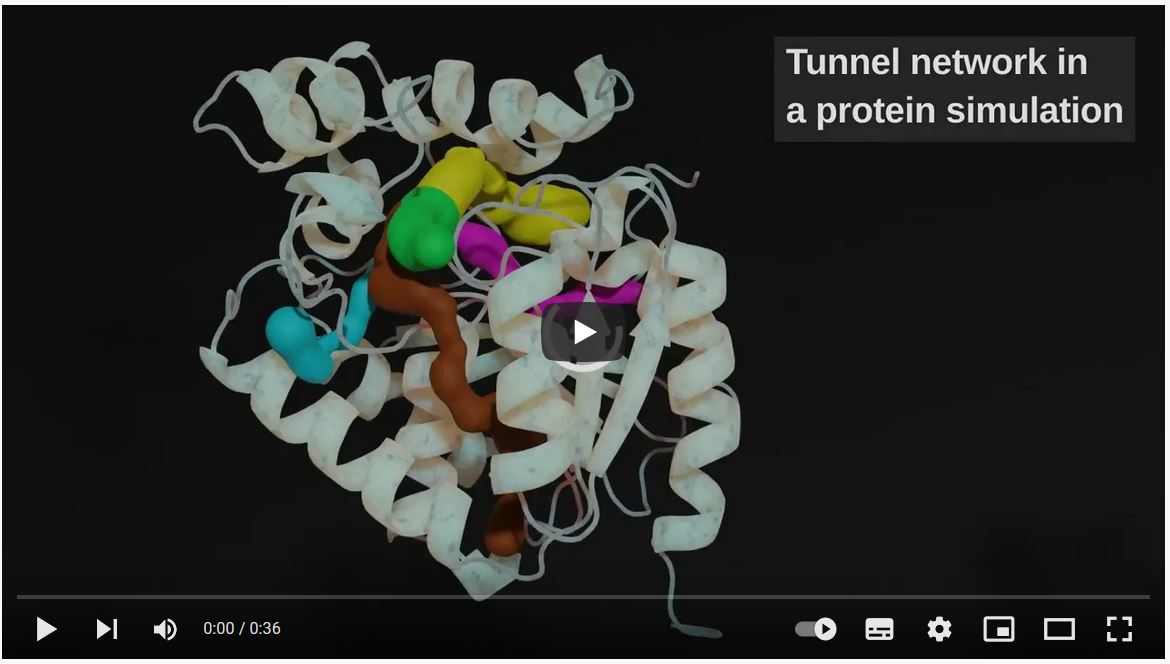
The transport of small organic molecules around the cell and beyond is critical for maintaining homeostasis. The transport is secured primarily by channels and tunnels created from the internal voids of biomolecules. As such, tunnels and channels represent critical targets for drug discovery and protein engineering projects. Unfortunately, these pathways are often equipped with dynamic gates, making them transient and difficult to study. One of the most effective approaches to characterize these rare events of ligand migration via transiently open pathways is to run molecular dynamics (MD) simulations. It is not uncommon that thousands of simulations need to be performed in order to catch the rare event when the pathway opens up and allows the ligand to pass. However, such high-throughput approaches impose a substantial burden on researchers in establishing the identity of the pathways observed across all simulations, determining which pathways are used by particular ligands, and developing means of specific quantitative analyses.
To this end, we present TransportTools: a library designed to alleviate these difficulties by providing easy and efficient access to comprehensive details on transport processes, even for large-scale simulation sets, and offering an environment for developing novel analyses and tools.
This library is helpful in case you would like to:
-
analyze transport pathways across extensive MD simulations, including those originating from massively parallel calculations or very long simulations;
-
integrate data on transport pathways and their actual utilization by small molecules again, possibly from a massive set of simulations;
-
compare transport processes under different settings, e.g., by contrasting transport in an original system against the same system perturbed by mutations, different solvents, or bound ligands.
You can have a glimpse of possible applications by following some of our three use-cases – data available on Zenodo:
use-case 1) discovering rare transient tunnels and their use by water molecules from 10 MD simulations
use-case 2) understanding of the effect of mutations on the system on transport pathways by contrasting simulation of wild-type with two mutants
use-case 3) determining the selectivity of pathways for substrate molecule from almost 600 MD simulations
TransportTools is implemented as a Python3 module and is available as pip and Conda packages. The source code and the user guide are available on GitHub.
The work has been published in Bioinformatics, 2021.