Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding – a new publication by the scientists from Laboratory of Protein Structure. Scientists from the Laboratory of Protein Structure published an article in Nucleic Acids Research which describes the structural insights in the mechanism of non-specific association of phosphorothioate-modified therapeutic nucleic acids with proteins. The work was performed in cooperation with Ionis Pharmaceuticals – a leading company in RNA-targeted therapeutics and Diamond synchrotron.
The phosphorothioate (PS) backbone is the most widely used modification in therapeutic nucleic acids, including antisense oligonucleotides (ASO). This modification involves a replacement of one of the two oxygen atoms in the repeating phosphate groups of the DNA with sulfur. PS-modified nucleic acids show improved properties, such as metabolic stability from nuclease-mediated degradation. One of the hallmarks of these compounds is enhanced interactions with cellular proteins. This property, on the one hand, facilitates cellular uptake of nucleic acid drugs and their cell retention, while, on the other hand, might contribute to cytotoxic properties of the drug molecule. Importantly, molecular mechanisms of interactions between PS nucleic acids and proteins have not been fully established.
To better understand how PS ASOs interact with cellular proteins, Hyjek-Składanowska et al. solved crystal structures of PS ASO bound to annexin A2 (AnxA2), a protein without any canonical DNA-binding domains, but previously implicated in the release of PS ASOs from endo-lysosomal compartments. In their work, the authors found that interactions between the sulfur atom of the PS linkage and the protein surface are mediated via lysine and arginine side chains and are mainly of hydrophobic character, suggesting that the hydrophobic nature of sulfur contributes to the association of PS ASOs with cellular proteins. Importantly, high quality of the crystals allowed the use of a unique experimental setup at the Diamond synchrotron to perform long-wavelength X-ray diffraction experiments. These experiments led to precise localization of the sulfur atoms in the structure and allowed the authors to establish which of the oxygen atoms are replaced with sulfur in the DNA bound to the protein. Interestingly, these results showed that stereoisomer preference at given phosphorothioate group in the DNA oligonucleotide is determined by the hydrophobic environment around the PS linkage coming not only from the protein but also from adjacent structural features within the DNA drug such as methyl groups on cytosine nucleobases. These studies will be very helpful in the design of a new generation of DNA-based drugs which will be stable and less toxic.
Malwina Hyjek-Składanowska, Brooke A Anderson, Vitaliy Mykhaylyk, Christian Orr, Armin Wagner, Jarosław T Poznański, Krzysztof Skowronek, Punit Seth, Marcin Nowotny, Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding, Nucleic Acids Research, 2022;, gkac774, https://doi.org/10.1093/nar/gkac774
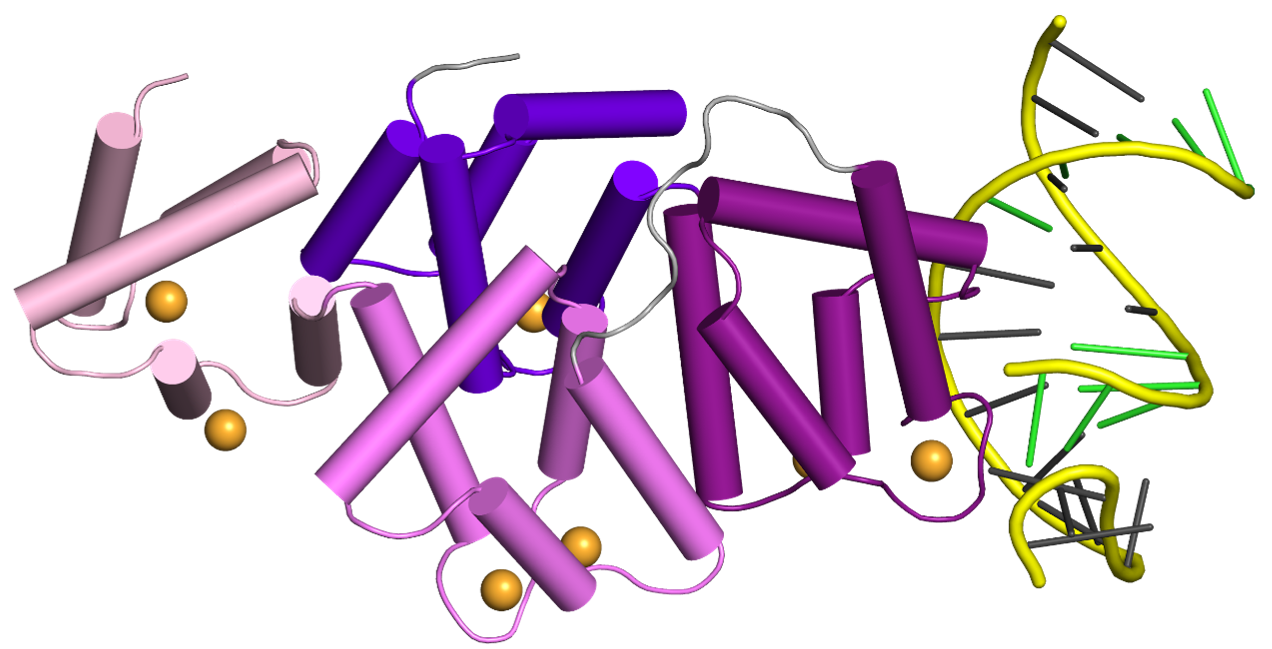
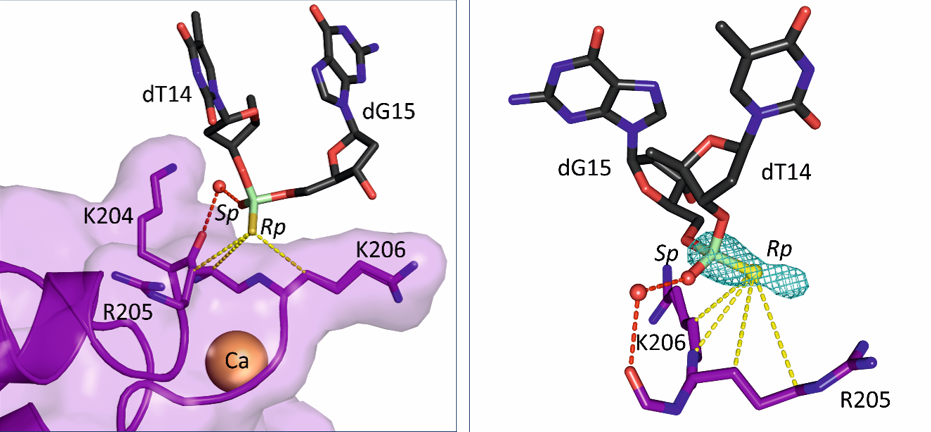
Crystal structure of AnxA2 in complex with PS ASO duplex. The protein is shown in purple and the therapeutic DNA in yellow. The middle panel shows the close-up on the phosphorothioate-binding surface in AnxA2. The right panel shows difference Fourier anomalous map calculated based on long-wavelength X-ray diffraction data (λ = 2.7552 Å, green mesh). The position of the sulfur-modified DNA bound by the proteins is determined by the hydrophobic interactions with surrounding amino acids.
