What do we know about the positive genetic memory or transcriptional memory? What does it depend on? What exactly are the factors responsible for it? A recent article published in Cellular and Molecular Life Sciences by researchers from the Laboratory of Structural Biology contributes to a better understanding of positive genetic memory.
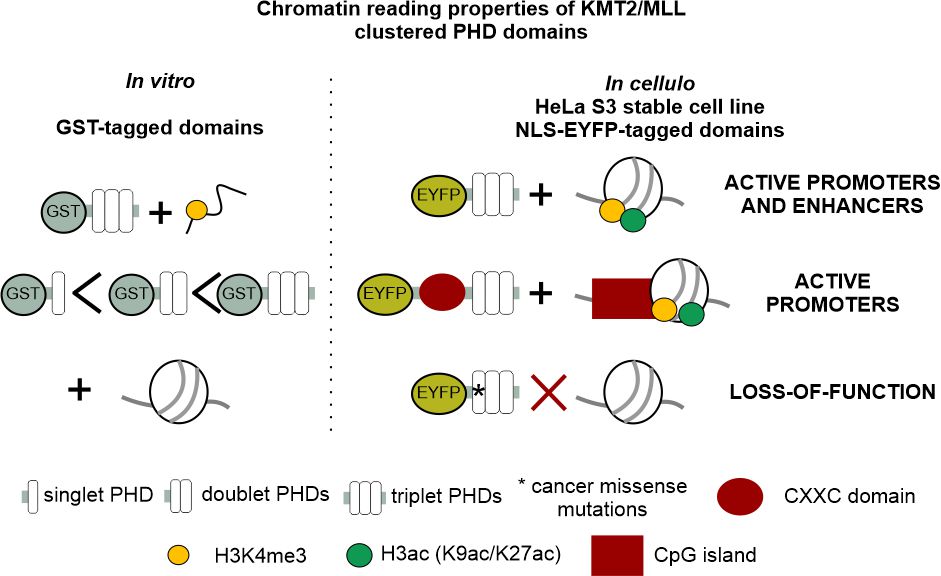
Genetic memory depends on a feed-forward loop that deposits H3K4 methylation, an activating chromatin mark, on already active promoters and enhancers. How the methyltransferases KMT2/MLL find their chromatin targets has not been understood. Stroynowska-Czerwinska and colleagues now show that clustered PHD domains in the KMT2/MLL methyltransferases alone are sufficient for the targeting. The known promoter or enhancer preferences of individual MLL/KMT2 methyltransferases are shown by the authors to be attributable to the presence or absence of the CXXC domains, respectively. The work presents the first genome-wide characterization of a histone reader domain in cells. MLL/KMT2 genes mutations are strongly associated with cancer. The authors show that even a single cancer-related missense mutation in the PHD domains can abolish targeting specificity. Therefore, the work also has medical implications.
Article reference:
Stroynowska-Czerwinska, A.M., Klimczak, M., Pastor, M. et al. Clustered PHD domains in KMT2/MLL proteins are attracted by H3K4me3 and H3 acetylation-rich active promoters and enhancers. Cell. Mol. Life Sci. 80, 23 (2023). https://doi.org/10.1007/s00018-022-04651-1
https://link.springer.com/article/10.1007/s00018-022-04651-1