It is our great pleasure to present the Annual Report 2022 of the International Institute of Molecular and Cell Biology in Warsaw.
The team of IIMCB Directors, researchers and administrators were active participants of the Envisioning the research centres of the future Conference organized for the 10th EU-LIFE Anniversary in Lisbon, Portugal, June 6-7, 2023.
The Conference created a vibrant and interactive space for exchanging ideas and discussions about how to be better researchers, more creative, proactive, sustainable, compassionate, inclusive, equitable and happy. It gathered scientific community, including researchers and scientific leaders, science administrators and policy makers, to dream up the ideal research places of the future.
During panel discussions Prof. Andrzej Dziembowski, Head of Laboratory of RNA Biology-ERA Chairs Group talked about the role of creativity in science, whereas Prof. Janusz Bujnicki, Head of Laboratory of Bioinformatics and Protein Engineering, gave a talk on how to be productive in the research world. Dr. Małgorzata Figiel provided conclusions to the audience from the discussions on creativity. IIMCB also presented two posters: “Centre of Excellence in RNA and Cell Biology – IIMCB of the future” co-authored by Dr. Iwona Pilecka, Dorota Libiszowska, Marcin Ogonowski and Alexia Danylow, and „Scientific excellence has no gender: IIMCB’s activities on gender equality, diversity & inclusion”, co-authored by Agnieszka Faliszewska, Katarzyna Fiedorowicz and Dr. Urszula-Białek Wyrzykowska.
The conference was preceded by the EU-LIFE Community Meeting. It took place at the hosting Instituto Gulbenkian de Ciência in Oeiras, where representatives of EU-LIFE institutes met to discuss issues in particular Working Groups.
EU-LIFE is an alliance of independent research institutes whose mission is to support and strengthen European research excellence. EU-LIFE members include leading research institutes that are internationally renowned for producing excellent research, widely transferring knowledge, and nurturing talent. Since its founding in 2013, EU-LIFE has become a stakeholder in science policy development, participating regularly in the European science policy dialogue. IIMCB became the first Polish member of the EULIFE alliance in 2020. As a member of EU-LIFE, IIMCB is working with 14 other institutes toward achieving and maintaining excellence in the life sciences, emphasizing quality and responsible science and highlighting issues that are related to European science policies.
The International Institute of Molecular and Cell Biology in Warsaw announces a recruitment for the positions of Intellectual Property Manager and Technology Transfer Manager. We invite you to take a look at the offer.
Intellectual Property Manager
The International Institute of Molecular and Cell Biology in Warsaw (IIMCB), Poland
invites applications for the position of Intellectual Property Manager
Are you passionate about knowledge valorization and are you ready to make a meaningful impact by translating IIMCB research discoveries into real-world applications? Are you open to expanding your expertise in intellectual property interests by engaging in a training-in-residence at the Flanders Institute for Biotechnology (VIB) in Belgium, our strategic partner? Are you ready to work in a dynamic team taking on the challenges of IP protection and technology transfer on an international level? If yes, please apply.
The IIMCB hosts a vibrant, multinational community of scientists and is well connected internationally as exemplified by the composition of the IIMCB's International Advisory Board and the IIMCB's membership in EU-LIFE, an alliance of 15 top European research institutions. The IIMCB is on its way to unprecedented scientific and institutional growth. Through expanding by 2025 to 20 research groups involved in innovative translational projects in RNA biology and cell biology, we aim to become a unique Polish center where excellent science is combined with professional IPR protection and commercialization. To this end, the IIMCB is establishing institutional support for translating basic science discoveries and research services into commercial applications and clinical innovations. The Intellectual Property Manager will be one of the key people in these processes. The successful candidate will focus on managing the intellectual property assets of the IIMCB by providing professional support in the identification, drafting of patent applications and licensing of IIMCB inventions. They will become a core staff of a dedicated technology transfer unit at the IIMCB and will benefit from a one-year training-in-residence at the VIB located in Ghent, Belgium, where they will gain expertise in identifying and processing intellectual property.
The establishment of a comprehensive system of technology transfer support is a part of the IIMCB’s institutional project entitled “RNA and Cell Biology - from Fundamental Research to Therapies” (RACE) funded in the Teaming for Excellence programme under Horizon Europe.
Key responsibilities:
- Creating an environment at the IIMCB that fosters innovation and entrepreneurial thinking
- Engaging with researchers to inspire and motivate them to think about the relevance of their research to society and the practical application of their discoveries
- Monitoring the results of research conducted in the IIMCB laboratories to determine their commercialization potential and recommending whether intellectual property protection should be sought
- Carrying out analyses of costs associated with patent prosecution
- Conducting patenting procedures (searching, drafting and prosecution)
- Organizing, managing and controlling all patents, trademarks and copyright files
- Overseeing IP related issues in externally funded research projects, including drafting IP agreements and managing IP during projects implementation
- Providing IP training to IIMCB researchers and advice on all IP-related matters
Necessary qualifications:
- PhD in life sciences
- Experience in biomedical research
- Hands-on knowledge of patent procedures is an asset
- Analytical mind, capable of analyzing a broad spectrum of life science topics and with the ability to deep-dive into scientific content
- Readiness for a 1-year training-in-residence at VIB, Ghent, Belgium
- High multitasking capacity combined with the ability to respect deadlines
- Excellent interpersonal and communication skills
- Fluency in oral and written Polish and English
The IIMCB offers:
- Employment contract with the IIMCB and secondment for 1-year training-in-residence at VIB, Ghent, Belgium
- Full-time salary in the range of 9.000 - 11.000 PLN gross, including the period of secondment, and an additional allowance for a secondment of 10.400 PLN gross
- Full-time salary is complemented by an additional 13th salary* plus an annual bonus.
- Attractive social package (co-financing Multisport cards, Christmas, Easter and summer holiday bonuses, etc.)
- Additional paid vacation time starting from the 2nd year of employment
- Work in hybrid mode
- Professional development and business travel opportunities
- Full organizational and administrative support by professional English-speaking staff
- Friendly, inclusive, international working environment
The application should include:
- Cover letter explaining the motivation for joining the IIMCB
- Curriculum Vitae
- A concise statement of 1-3 professional achievements
- Contact information for two references
Applications should be sent to This email address is being protected from spambots. You need JavaScript enabled to view it., in the subject please include "IPM" and your first and last name.
The deadline for applications is June 15, 2023.
Job interviews are scheduled for June 22-23, 2023.
Applications should include the statement: “I hereby agree to the processing of my personal data, included in the application documents by the International Institute of Molecular and Cell Biology in Warsaw, 4 Księcia Trojdena Street, 02-109 Warsaw, for the purpose of carrying out the current recruitment process.” Your personal data will be processed only for the purpose of the recruitment procedure by the International Institute of Molecular and Cell Biology in Warsaw. Full information is available at https://bit.ly/3UFWpY2.
*13th salary is calculated as 8.5% of the gross salary received in a given calendar year (excluding sick pay) under the condition of having worked a minimum of 6 months in a given year.
Technology Transfer Manager
The International Institute of Molecular and Cell Biology in Warsaw (IIMCB), Poland
invites applications for the position of Technology Transfer Manager
Are you passionate about knowledge valorization and are you ready to make a meaningful impact by translating IIMCB research discoveries into real-world applications? Are you open to expanding your expertise by engaging in the training-in-residence at the Flanders Institute for Biotechnology (VIB) in Belgium, our strategic partner? Are you eager to build a dynamic team taking on the challenges of IP protection and technology transfer on an international level? If yes, please apply.
The IIMCB hosts a vibrant, multinational community of scientists and is well connected internationally as exemplified by the composition of the IIMCB's International Advisory Board and the IIMCB's membership in EU-LIFE, an alliance of 15 top European research institutions. The IIMCB is on its way to unprecedented scientific and institutional growth. By expanding to 20 research groups involved in innovative translational projects in RNA biology and cell biology by 2025, we aim to become a unique Polish center where excellent science is combined with professional IPR protection and commercialization. To this end, the IIMCB is establishing institutional support for translating basic science discoveries and research services into commercial applications and clinical innovations. The Technology Transfer Manager will be at the heart of these processes acting as an interface between the IIMCB scientists and potential clients, including clinics and businesses. They will become a core staff of the dedicated technology transfer unit at the IIMCB and will develop an array of activities for effective IIMCB knowledge valorization. The Technology Transfer Manager will benefit from 1.5-year training-in-residence at the Flanders Institute for Biotechnology (VIB) located in Ghent, Belgium where they will gain expertise in identifying intellectual property with commercialization potential, validating research discoveries, establishing partnerships with industry, and using various knowledge transfer models.
The establishment of a comprehensive system of technology transfer support is a part of the IIMCB’s institutional project entitled “RNA and Cell Biology - from Fundamental Research to Therapies” (RACE) funded in the Teaming for Excellence programme under Horizon Europe.
Key responsibilities:
- Creating an environment at the IIMCB that fosters innovation and entrepreneurial thinking
- Engaging with researchers to inspire and motivate them to think about the relevance of their research to society and the practical application of their discoveries
- Monitoring the results of research conducted in the IIMCB laboratories and ensuring proper protection of results with commercialization potential
- Seeking funding sources and business partners for the development of inventions
- Applying for grants for technology development and commercialization, implementing and reporting on them
- Building and presenting business cases and marketing materials with the aim of partnering with the biotech industry (R&D collaborations and/or licensing)
- Expanding the IIMCB's business network and building the IIMCB's image as a business partner
- Cooperating with lawyers and patent attorneys in preparation of legal documents and their follow up (alliance management)
- Monitoring IIMCB discoveries along the patenting and commercialization processes
Necessary qualifications:
- PhD in life sciences
- Experience in biomedical research
- Experience in translational projects and technology transfer is an asset
- Strong interest to develop professional competences in technology transfer
- Readiness for a 1.5-year training-in-residence at VIB, Ghent, Belgium
- Strong analytical skills
- Decision-making and negotiation skills
- Excellent interpersonal and communication skills
- Fluency in oral and written Polish and English
The IIMCB offers:
- Employment contract with the IIMCB and secondment for 1.5-year training-in-residence at VIB, Ghent, Belgium
- Full-time salary in the range of 12.000-14.000 PLN gross, including the period of secondment, and an additional allowance for secondment of 10.400 PLN gross
- Full-time salary is complemented by an additional 13th salary* plus an annual bonus
- Attractive social package (co-financing Multisport cards, Christmas, Easter and summer holiday bonuses, etc.)
- Additional paid vacation starting from the 2nd year of employment
- Work in hybrid mode
- Professional development and business travel opportunities
- Full organizational and administrative support by professional English-speaking staff
- Friendly, inclusive, international working environment
The application should include:
- Cover letter explaining the motivation for joining the IIMCB
- Curriculum Vitae
- A concise statement of 1-3 professional achievements
- Contact information for two references
Applications should be sent to This email address is being protected from spambots. You need JavaScript enabled to view it., in the subject please include "TTM" and your first and last name.
The deadline for applications is June 15, 2023.
Job interviews are scheduled for June 22-23, 2023.
Applications should include the statement: “I hereby agree to the processing of my personal data, included in the application documents by the International Institute of Molecular and Cell Biology in Warsaw, 4 Księcia Trojdena Street, 02-109 Warsaw, for the purpose of carrying out the current recruitment process.” Your personal data will be processed only for the purpose of the recruitment procedure by the International Institute of Molecular and Cell Biology in Warsaw. Full information is available at https://bit.ly/3UFWpY2.
*13th salary is calculated as 8.5% of the gross salary received in a given calendar year (excluding sick pay) under the condition of having worked a minimum of 6 months in a given year.
Dr. Aleksandra Anna Kołodziejczyk with a new NCN grant!
Dr. Aleksandra Kołodziejczyk, the Head of the Laboratory of Cellular Genomics at the International Institute of Molecular and Cell Biology in Warsaw, has received funding under the SONATA 18 scheme, organized by the National Science Centre (NCN). Dr. Kołodziejczyk has been awarded with a grant of 2 544 840 PLN for the project entitled: "The role of gut-liver axis in Amanita species mushroom poisoning". Dr. Kołodziejczyk took the first place within NZ5 panel projects in the ranking list. Congratulations!
The aim of the project is to gain comprehensive understanding of the molecular and cellular mechanisms underlying the pathophysiology of Amanita phalloides poisoning with a goal in mind to improve clinical outcomes. The description of the project is available here.
The project will last 36 months and it will be carried out by a consortium of two institutions, namely, the International Institute of Molecular and Cell Biology in Warsaw and Medical University of Warsaw.
Dr hab. Cecilia Winata - laureate of the OPUS 24 scheme!
Dr hab. Cecilia Winata, Head of the Laboratory of Zebrafish Developmental Genomics at the International Institute of Molecular and Cell Biology in Warsaw, has received funding under the OPUS 24 scheme, organized by the National Science Centre (NCN). Dr hab. Cecilia Winata has been awarded a grant of 3 040 240 PLN for the project entitled: "Elucidating the contribution of non-coding genomic elements to heart development and disease at single-cell resolution". Dr hab. Cecilia Winata took the first place within NZ2 panel projects in the ranking list. Congratulations!
The project will last 48 months. Its goal is to reveal novel congenital heart disease-causing genetic factors and mechanisms, and contribute critical insights into understanding the mechanism of non-coding genetic variants in the pathogenesis of this condition. The description of the project is available here.
On May 11, 2023, Professor Marta Miączyńska, the Director of the International Institute of Molecular and Cell Biology in Warsaw (IIMCB), signed a contract for the preparation of design documentation for Institute's new seat with an architectural studio Atelier Tektura Sp. z o.o, the winner of the competition for the best architectural concept for Institute's seat.
The signing of the contract is another step that brings us closer to the new seat, which is so important for Institute's further development. We are convinced that cooperation with Atelier Tektura will be successful and it will allow to achieve the set goal in the planned time – said Prof. Marta Miączyńska, the Director of the IIMCB.
In December 2022, the competition for the architectural concept of Institute's new seat was settled. The winner was Atelier Tektura. The winning project was recognized for its very good, rational, and logical solution of rooms layout. The flexibility of the project was also appreciated, namely, the possibility of modifying the layout of the space as well as the construction solutions.
4 scientific disciplines, 9 renowned research institutes, 1 unique and interdisciplinary training program - this is the characteristics of Warsaw PhD School in Natural and BioMedical Sciences. Its interdisciplinary PhD program offers cutting-edge research opportunities in the areas of physics, chemistry, biology, and medicine, as well as it provides the possibility of collaboration with leading research centers around the world. Exactly, on May 4, 2023, an international campaign to promote Doctoral School on social media was launched. Its goal is to present the School's full offer and to encourage potential PhD students to study in Warsaw. The campaign debuted with a promotional video which can be watched HERE. The recruitment starts on Monday, May 22. You can read about the details on the research projects you can apply in the text below.
Science is getting more and more interdisciplinary. To discover something truly new and groundbreaking, different research methods need to be used. Only the synergy of different scientific disciplines can lead to fascinating discoveries. This is exactly what Warsaw-4-PhD Doctoral School offers, hosting currently students from 25 different countries. Thanks to a constant and long-term cooperation with foreign research centers, PhD students have great opportunities for development, including the international cooperation. Obtaining a doctoral degree opens up the possibility of pursuing a research career in the most modern laboratories in the world.
Recruitment for the upcoming call will begin on May 22, 2023 and last until June 4. By clicking on the link below HERE, you can find the research projects which can be chosen by potential doctoral students within the current recruitment process.
The Warsaw PhD School in Natural and BioMedical Sciences consists of:
Nencki Institute of Experimental Biology PAS
Institute of Organic Chemistry PAS
Institute of Physical Chemistry PAS
Institute of Physics PAS
Center for Theoretical Physics PAS
Institute of High Pressure Physics PAS
Maria Sklodowska – Curie National Research Institute of Oncology
Institute of Psychiatry and Neurology
International Institute of Molecular and Cell Biology in Warsaw
More: https://warsaw4phd.eu/
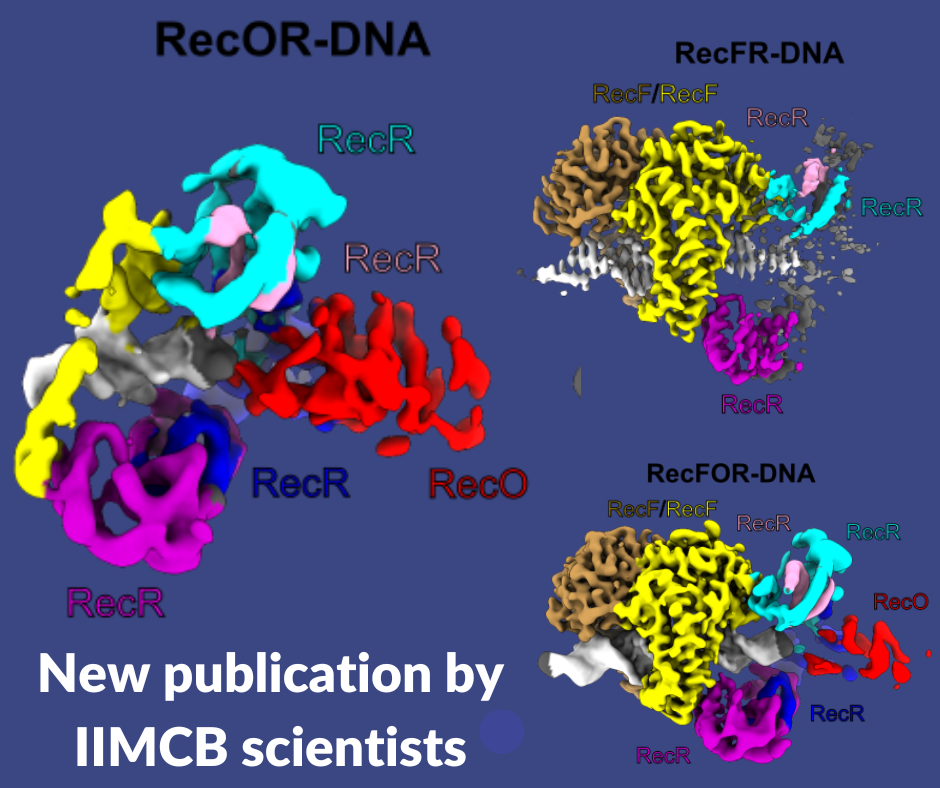
Scientists from the Laboratory of Protein Structure at the International Institute of Molecular and Cell Biology in Warsaw explained the mechanism of a key protein complex in bacterial homologous recombination.
Homologous recombination is one of the fundamental pathways for the repair of damaged genetic information. It involves the generation of single-stranded DNA at the site of damage. This DNA performs a homology-based search for the template. In bacteria, one of the homologous recombination-based repair pathways involves RecF, RecO and RecR proteins. They bind at the junction of single-stranded (ss) and double-stranded (ds) DNA and then facilitate the replacement of the SSB protein, which initially covers ssDNA, with RecA which promotes the search for the homologous sequence. However, the molecular mechanism of RecFOR cooperation in the pathway was largely unknown. Shivlee Nirwal and co-workers from Prof. Marcin Nowotny’s laboratory used the SOLARIS National Synchrotron Radiation Centre to determine the cryo-electron microscopy structures of the RecF-DNA and RecFOR-DNA complexes, and their subcomplexes, to elucidate the mechanism of the cooperation of these proteins in ss-dsDNA junction recognition.
The RecF-DNA and RecFR-DNA subcomplex of the RecFOR-DNA assembly, show how the RecF dimer uses helical protrusions in its structure to bind double-stranded DNA. The RecFR-DNA subcomplex further shows the way one RecF protomer interacts with two different regions of a ring made of four RecR molecules which stabilize this ring on DNA. The lower-resolution reconstructions of the RecR–RecO subcomplex and the RecFOR–DNA assembly explain how RecO is positioned to interact with ssDNA and SSB, which is proposed to lock the complex on a ssDNA–dsDNA junction. This mode of RecFOR action is further supported by biochemical and biophysical experiments. Overall, the results obtained integrate and explain the wealth of biochemical data available for the RecFOR system and provide a framework for a complete understanding of the RecFOR homologous recombination pathway.
Link to the publication: https://www.nature.com/articles/s41594-023-00967-z
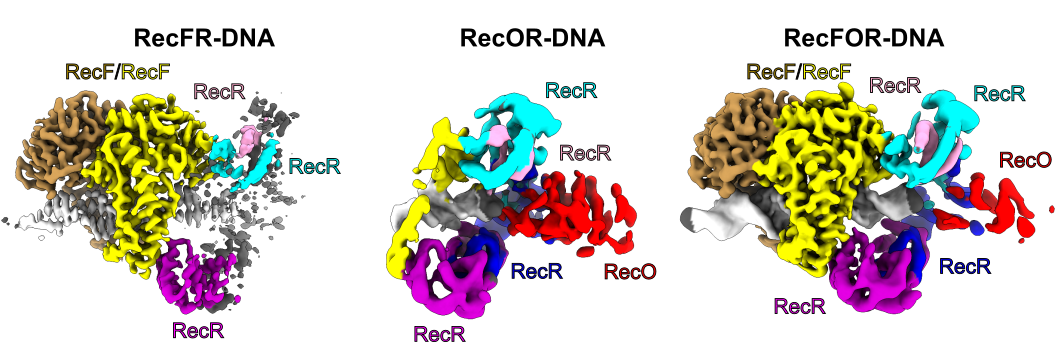
Fig: The cryo-EM reconstructions for the RecFOR-DNA assembly and the RecFR-DNA and RecOR-DNA subcomplexes of the assembly.
A two-month internship in one of the IIMCB's well-equipped 14 laboratories, under the guidance of outstanding scientists and experts in their fields – this is an offer of the internship program introduced by the International Institute of Molecular and Cell Biology in Warsaw.
The offer is aimed at talented life sciences students interested in the field of biology and biotechnology. During the internship, selected students will have the opportunity to participate in research projects being currently carried out in the IIMCB laboratories, under the supervision of excellent scientists. It will also be an excellent opportunity to work in international teams on issues related to the development of modern science as well as an opportunity to improve own scientific and analytical skills.
Students whose applications will be approved, will be offered a one-month full-time internship with a salary in the amount of PLN 2.500 net/month. The IIMCB will provide them with initial scientific training and guidelines from the experienced scientists and PhD students. In addition, the apprentices will obtain full organizational and administrative support in a friendly and international work environment.
Students interested in the offer are encouraged to learn more about the research topics of individual laboratories and send their application (CV and registration form) to: This email address is being protected from spambots. You need JavaScript enabled to view it.
The applications for the internship program are accepted on a rolling basis. Undergraduate students, graduate students, and recent graduates are invited to send their applications.
The number of places is limited.
We are pleased to announce that as of the 1 April two new Laboratories started their activities at the International Institute of Molecular and Cell Biology in Warsaw: Laboratory of Single-Molecule Biophysics, headed by Dr. Ewelina Małecka-Grajek and Laboratory of Cellular Genomics, headed by Dr. Aleksandra Kołodziejczyk.
In the Laboratory of Single-Molecule Biophysics the researchers are focused on understanding the coordination between RNA targeting, degradation, and translation in bacteria. Most species of bacteria are harmless and are often beneficial, but others can cause infectious diseases. The Laboratory also performs research using the TIRF microscopy method which allows the visualization of hundreds of biomolecules immobilized on the microscopic slide. However, each molecule is analyzed separately.
In the Laboratory of Cellular Genomics the main research area is investigating the bidirectional crosstalk between the liver and the intestine, focusing on the role of resident microbiota. The researchers also explore how shifts in bacterial composition and changes in intestinal physiology affect liver cells and how liver health contributes to the homeostasis in the gut.They are also focused on the mechanisms underlying chronic inflammatory processes accompanying organ fibrosis, metabolic syndrome and autoimmune disorders. The scientists are also exploring the role of microbiota-derived metabolites in the host physiology, with special regard to the the molecular mechanisms by which metabolites affect cellular functions.