-
May 25, 2020 - recruitment opens
-
June 7, 2020 - deadline for applications
Research projects for admissions 2020/2021-1:
Linking abnormal Ca2+ signaling and the unfolded protein response with Huntington’s disease pathology in both YAC128 mouse model and iPSC-derived neurons from HD patients.
Prof. Jacek Kuźnicki, PhD; Magdalena Czeredys, PhD, Laboratory of Neurodegeneration
Description: Huntington’s disease (HD) is a progressive neurodegenerative disorder characterized by the aggregation of mutant huntingtin and degeneration of medium spiny neurons (MSNs) in the striatum. Abnormal Ca2+ signaling is considered as an early event in HD pathology since disturbances in Ca2+ homeostasis were found in HD models and postmortem samples from HD patients. One of the pathways for Ca2+ signaling is store-operated calcium entry (SOCE). The activation of inositol-(1,4,5)triphosphate receptor 1 (IP3R1) results in Ca2+ release, which decreases ER Ca2+ content and activates Ca2+ influx through SOC channels. Elevated SOCE and increased IP3R1 activity was previously reported in MSNs from the transgenic model of HD, YAC128. The project is based on the hypothesis that neurodegeneration in HD is induced by disturbances in Ca2+ signaling in neurons. Previously we found that huntingtin-associated protein 1 (HAP1) that is overexpressed in striatal neurons and binds to mutant huntingtin causes dysregulation of Ca2+ signaling by increased activation of both SOCE and IP3R1 receptors. We intend to examine the link between dysregulated Ca2+ signals and neuronal cell death in HD. The experiments will be performed in YAC128 MSNs cultures and neurons delivered by the reprogramming of fibroblasts from HD patients with the application on CRISPR/Cas9-based editing strategies and Ca2+ signaling inhibitors.
Aim: The project aims to investigate whether and how the disturbed Ca2+ homeostasis affects HD pathology. A Ph.D. project related to this issue will be done using different HD models. One position is available in the project. We are looking for a person interested in neurobiology, with experience in working with animal models (mice, zebrafish), cell cultures, and biochemical techniques (immunoprecipitation, western blot). Knowledge/experience in iPSCs cultures is welcome.
Number of positions available: 1
Source of funding: NCN/OPUS grant
Contact: This email address is being protected from spambots. You need JavaScript enabled to view it.
Cytoplasmic polyadenylation as a central regulator of physiological processes
Prof. Andrzej Dziembowski, PhD, Laboratory of RNA Biology
Description: Poly(A) tails generated by canonical poly(A) polymerases during mRNA 3’ formation are essential for mRNA stability and translation. It is now appreciated that poly(A) tail dynamics is more complex than previously suspected; deadenylated mRNAs in the cytoplasm can be degraded, uridylated or stored in a dormant state to be later re adenylated to activate protein synthesis. Cytoplasmic polyadenylation was mostly studied in the context of gametogenesis and in synapses, where the transcriptional activity is limited. Surprisingly, mouse lines devoid of the well known cytoplasmic poly(A) polymerase GLD2 display no apparent phenotypes. We recently described a novel family of cytoplasmic poly(A) polymerases, TENT5 (FAM46), comprising four members in vertebrates (Mroczek et al. & Bilska et al. Nat Comm 2017,2020). TENT5C acts as a tumor suppressor in multiple myeloma, while mutations in TENT5A lead to a rare disease osteogenesis imperfecta. We have generated KO mouse models for all TENT5 genes and detected a variety of different phenotypes affecting several organs and biological processes: gametogenesis, growth, skeletal development, hematopoiesis, immune response, and behavior. Moreover, analysis of the KO of worm TENT5 orthologue revealed dysfunction of innate immunity. Thus, TENT5 proteins contribute significantly to metazoan physiology and, more generally, that cytoplasmic polyadenylation plays a much broader role than previously anticipated, opening a new area of important research.
Aim: The project aims is to dissect functions and mechanisms of cytoplasmic polyadenylation by TENT5 in innate immunity, erythropoiesis, and neuronal physiology. Unique animal models constructed using CRISPR/Cas9, combined with advanced transcriptomic and proteomic approaches, will be used to achieve our goals. Several positions are available in the lab. The exact PhD project will depend on the particular skills and preferences of the student. We are looking for students with experience in work with animal modes (mouse, C. elegans), RNA biology, or bioinformatics.
Number of positions available: 4
Source of funding: NCN/Norway grants/FNP/Horizon2020 era chairs
Contact: This email address is being protected from spambots. You need JavaScript enabled to view it.
See also:
The International Institute of Molecular and Cell Biology in Warsaw (IIMCB) educates PhD students within the framework of the Warsaw PhD School in Natural and BioMedical Sciences (Warsaw-4-PhD) and conducts the procedure for awarding the doctoral degree.
|
The Warsaw-4-PhD School was established by nine research institutions in Warsaw and offers doctoral education in four disciplines: biology, chemistry, physics, and medical sciences. Candidates apply to specific research projects carried out within one of the participating institutions. |
PhD at IIMCB
- We offer the opportunity to carry out research projects in RNA and cell biology, as well as to complete the entire PhD procedure directly at IIMCB – from education to the dissertation defense.
- Recruitment is held three times a year in the form of an open international competition. Studies begin in the winter or summer semester.
- Each PhD student carries out an individual research plan under the supervision of a mentor. A mid-term evaluation is conducted halfway through the program.
Who is eligible?
- Candidates holding a Master’s degree or equivalent,
- Interested in conducting research in the biological sciences, and
- Proficient in English to a level that enables work in an international research environment.
What do we offer?
- Free education at the doctoral school.
- A scholarship of PLN 5,500 before the mid-term evaluation and PLN 6,000 after the evaluation, plus support from the Grants Office in applying for additional funding.
- Academic supervision and mentoring – close cooperation with a supervisor and support from a research team.
- A highly qualified scientific staff conducting research published in renowned international journals.
- An international working environment with English as the main language of communication.
- Comprehensive support for foreign students – we assist with obtaining a visa, legalizing residence, and completing all other formalities.
- Opportunities for international development through participation in conferences, workshops, foreign internships, with financial support for travel.
- Administrative assistance from the PhD Office.
- Health and social benefits – co-financing for private medical care, sports cards, training, cultural events, and other benefits available for IIMCB employees.
- Participation in institutional dialogue – PhD student representatives take part in regular meetings with the Institute’s management and the International Advisory Board.
How to apply?
- The next recruitment will begin on December 18, 2025, and will last until January 6, 2026. At that time, information on available research projects will be posted in the news section of the Institute’s website.
- More details about the application process can be found on the Warsaw-4-PhD School website: https://warsaw4phd.eu
Contact
Agnieszka Faliszewska
Deputy Head of HR Department
This email address is being protected from spambots. You need JavaScript enabled to view it.

Warsaw PhD School of Natural and BioMedical Sciences [Warsaw-4-PhD] was established on May 16, 2019. The School is operated by nine research institutions: 1. Nencki Institute of Experimental Biology PAS (leader of the School); 2.Institute of Organic Chemistry PAS; 3. Institute of Physical Chemistry PAS; 4. Institute of Physics PAS; 5.Center for Theoretical Physics PAS; 6. Institute of High Pressure Physics PAS; 7. Maria Sklodowska-Curie Institute - Oncology Center; 8. Institute of Psychiatry and Neurology; 9. International Institute of Molecular and Cell Biology in Warsaw. The School educates doctoral students in four scientific disciplines: biology, chemistry, physics and medicine.The School commenced its activity on October 1, 2019.
IIMCB contact:
Zuzanna Mackiewicz (Laboratory of RNA Biology)
e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
Human Resources Unit
e-mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
International Institute of Molecular and Cell Biology in Warsaw
4 Ks. Trojdena Street, 02-109 Warsaw, Poland

eduroam®
Visitors from eduroam-affiliated institutions may use the eduroam network that is now available at IIMCB in Warsaw.
There is no need for additional configuration of your mobile device if it was provisioned at your primary institution also being a member of the eduroam alliance.
Legal and regulatory information: Wyciąg z regulaminu Eduroam / Excerpt from Eduroam terms of usage
Wi-Fi Range:
- eduroam Wi-Fi is available on-site on all floors of the Institute building
Connection parameters:
- Network name: eduroam
- Credentials: Single-Sign-On: <username>@iimcb.gov.pl> or similar for visiting guests
- Encryption: WPA2-Enterprise (802.1x)
- Cipher: CCMP
- Authentication: TTLS+MSCHAPv2 / PEAP+MSCHAPv2
- Client certificates: not required (for IIMCB users)
- Server certificates: preinstalled (for IIMCB users) / do not validate (*1),
- or add our certificates to your trust keystore: https://ftp.users.genesilico.pl/eduroam/ )
Installers:

https://cat.eduroam.org/?idp=6012
Configuration instructions:
To set up the connection manually, follow these instructions:
- Android (PL) / Android (EN) (connection profile)
- Windows
- Linux
(*1) - until tested further

Project number: 2015/19/P/NZ2/03278
Project title: “"Deciphering BMP6 regulatory mechanisms using CRISPR/Cas9-based screening approach”
Project leader: dr. Katarzyna Mleczko-Sanecka
Source of funding: National Science Centre, Poland
Budget: 893 104 PLN
Project Duration: 01.01.2017-31.12.2018
About the project:
Maintenance of an appropriate iron balance is essential for the correct functioning of the organism, and thus an increasing knowledge of the genetic control of iron homeostasis is important for human health. To prevent iron-overload diseases, excessive iron levels must be correctly sensed to stimulate expression of the iron-regulatory hormone hepcidin that limits further iron absorption from the diet. The objective of our work is to better understand the process of iron-sensing in the liver. We are seeking to identify still elusive mechanisms that control iron-triggered induction of bone morphogenetic protein 6 (BMP6), a key iron-sensing cytokine produced by the liver sinusoidal endothelial cells (LSECs) that stimulates hepcidin production. Our preliminary data suggest that a factor secreted by iron-loaded hepatocytes rather than iron deposition in LSECs serves as a signal to enhance endothelial BMP6 expression. Using cell lines and primary liver cell cultures, we are now characterizing one candidate protein that may mediate communication between iron-laden hepatocytes and LSECs. We expect that our work which will shed light on a novel iron-sensing mechanism in the liver, that leads to induction of the BMP6-hepcidin axis.
Publications:
Mleczko-Sanecka K, da Silva AR, Call D, Neves J, Schmeer N, Damm G, Seehofer D and Muckenthaler MU. Imatinib and spironolactone suppress hepcidin expression Haematologica. 2017 Jul;102(7):1173-1184.
Posters at the international conferences:
Poster at the Seventh Congress of the International BioIron Society (IBIS) Biennial World Meeting (BioIron 2017), May 7 – 11, 2017, UCLA, Los Angeles, USA. IMATINIB AND SPIRONOLACTONE SUPPRESS HEPCIDIN EXPRESSION. Katarzyna Mleczko-Sanecka, Debora Call, Ana Rita da Silva, Nikolai Schmeer, Melanie Kiessig, Georg Damm and Martina Muckenthaler. Presented By: Katarzyna Mleczko-Sanecka, PhD
“MOlecular Signaling in Health and Disease - Interdisciplinary Centre of Excellence”
MOSaIC
H2020 WIDESPREAD-03-2017: ERA Chairs - 810425
BASIC FACTS
Project title: “MOlecular Signaling in Health and Disease - Interdisciplinary Centre of Excellence”
Project acronym: MOSaIC
Project implementation period: 1 November 2018 - 31 October 2023
Granted amount: 2 498 887,50 €
Referenced call: H2020 WIDESPREAD-03-2017
Project coordinator: Prof. Jacek Kuźnicki
NEWS
We will keep you updated on MOSaiC activities. Please follow the project’s news at:



CONCEPT AND OBJECTIVES
Creation of the Interdisciplinary Centre of Excellence in Molecular Signaling in Health and Disease (MOSaIC)
The ERA Chairs scheme gives us a unique opportunity to improve our performance by attracting an outstanding principal investigator in molecular signaling. This investigator will complement and strengthen the current research lines at IIMCB and will influence structural changes for responsible research and effective science management.
The MOSaIC has 5 objectives. The first is recruitment of an ERA Chair holder and affiliates in an open process that involves a set of incentives for the successful candidate to establish an excellent research group. The second is to augment scientific research and innovation in IIMCB and surrounding region through the work of the ERA Chair holder in molecular signaling, a field with great promise for medical advancement. The third and fourth involve structural improvements in science management and consolidated HR activities to develop a research environment that meets the highest international standards, including those set in the EU Charter and Code. Dissemination and communication activities will enable us to achieve the fifth objective: widespread recognition of the MOSaIC and IIMCB among national and European stakeholders.
Thanks to the MOSaIC, IIMCB scientists will lead innovative and internationally competitive research in an environment of excellent organizational standards and support. They will engage in European partnerships, and their projects will lead to practical inventions.
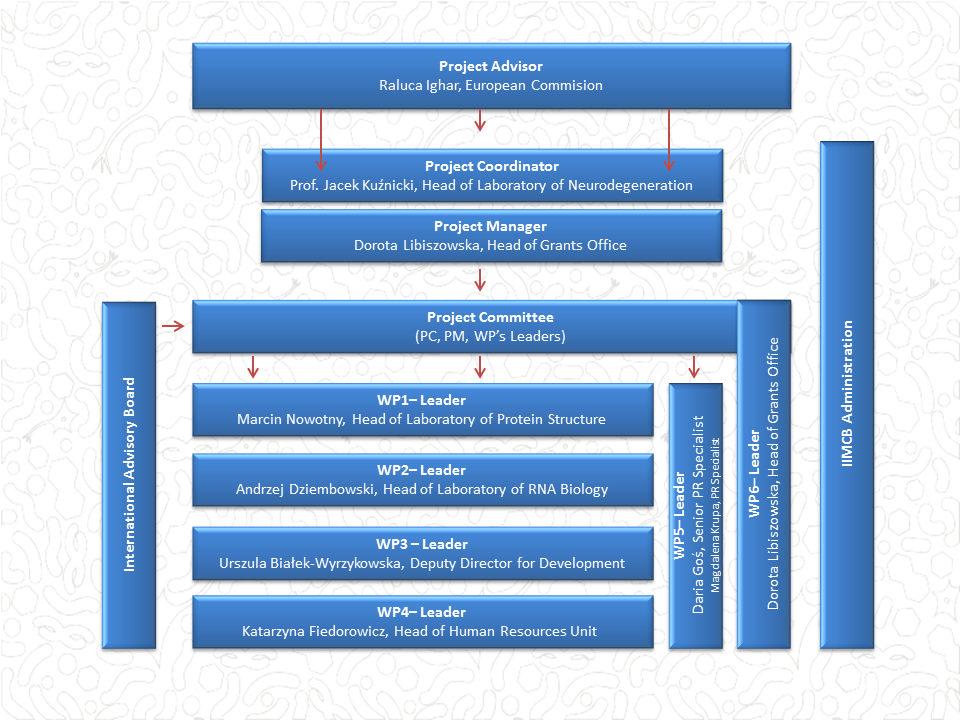
WORK PLAN
Project coordinator: Jacek Kuźnicki, IIMCB Director
WP 1. Recruitment of the ERA Chair holder and new group members
Leader: Marcin Nowotny, Head of Laboratory of Protein Structure
We will recruit an outstanding mid-career scientist with proven leadership experience for a Senior Group Leader position. The ERA Chair holder will be provided with the best conditions for research, including optimal laboratory and office space, free access to equipment, and additional benefits. Once recruited, the new Group Leader will autonomously build his or her own research group. The above activities will be coordinated by Marcin Nowotny, Head of the Laboratory of Protein Structure. The recruitment process of the ERA Chair holder will involve active participation of IIMCB International Advisory Board and the Head of HR Unit. The latter will also thoroughly support the ERA Chair holder in the recruitment of new laboratory members.
WP 2. Strengthening scientific and innovation potential of IIMCB and the surrounding region
Leader: ERA Chair holder
To boost IIMCB research and innovation, increase its competitiveness, and stay connected with partners across Europe, the ERA Chair holder and their group will generate excellent results of their own research in the field of molecular signalling in health and disease and to contribute to the overall position of IIMCB in the scientific, local, regional, and European landscape. For the scientific functioning of the new group (especially in the initial phase), the ERA Chair Holder will be mentored by Dr. Nowotny. All formal, administrative, and financial arrangements will be assisted by the corresponding administrative units: the Grants Office, Scientific Coordination, Accounting, HR, public relations, and IT.
WP 3. Structural changes for effective science management at IIMCB
Leader: Urszula Bialek-Wyrzykowska, Deputy Director for Development
To support research and innovation at IIMCB, we will improve specific internal policies by expanding and/or adopting them to national and European recommendations, including ERA priorities. The proposed structural changes cover 3 distinct areas in good scientific practices (OA, ODM, ethics) and IPR management at the Institute. They fall under responsibility of the Deputy Director for Development, Urszula Bialek-Wyrzykowska, who will be the WP3 Leader. She will be assisted by dedicated personnel from the Scientific Coordination Unit and the Institute’s IPR manager. A key element will be conceptual input and advice on the implementation of WP3 tasks from the ERA Chair holder.
WP4. Structural changes for more effective implementation of C&C principles
Leader: Katarzyna Fiedorowicz, Head of Human Resources Unit
To implement the EU Charter & Code principles in a comprehensive way and to respond to our scientists’ needs, we will enhance the functioning of the HR Unit to give a professional rank to EU C&C-related activities and to develop them systematically. These include elaboration of the HR Strategy for IIMCB, assisting scientists in recruitment processes, organization of 5 career development training sessions for IIMCB employee groups, support to foreign employees during their stay in Poland, efficient conflict solving and coordination of gender issues. For effective implementation of these ambitious assignments, before the start date of MOSaIC, we recruited an experienced, full-time Head of HR Unit, Ms. Katarzyna Fiedorowicz, who assumes the role of WP4 Leader. Ms. Fiedorowicz will be engaged in recruitment of the ERA Chair holder and group members.
WP5. Strengthening MOSaIC’s recognition among national and European stakeholders.
Leader: Daria Gos, PR Manager
Dissemination, application, and communication activities are indispensable elements that maximize the impact of MOSaIC’s achievements. Along with RRI principles, we will explore various means of dissemination and communication and will target numerous communities. The tasks assigned to WP5 will be overseen by experienced PR Manager, Daria Gos. WP5 tasks include Dissemination of MOSaIC results by the ERA Chair holder and group members to scientists and potential business partners, dissemination of results of structural improvements, communicating MOSaIC and its results to different stakeholders and elaboration of promotional tools and materials.
WP6. Management
Leader: Dorota Libiszowska, Head of Grants Office
The work involves high-level coordination and management of the project, including monitoring of work progress and assessment of the implementation of distinct WPs. Project monitoring will be done by the Project Committee that consists of the Project Coordinator and WP Leaders. Project Committee will meet annually to monitor work progress, discuss strategic steps, and respond to unexpected situations. These meetings will involve members of the International Advisory Board who will assess how the project helps achieve the strategic goals set up for IIMCB and will formulate recommendations for further development. IAB will play a crucial role in the selection process of the ERA Chair holder, it will assess the scientific achievements and progress of the new group.

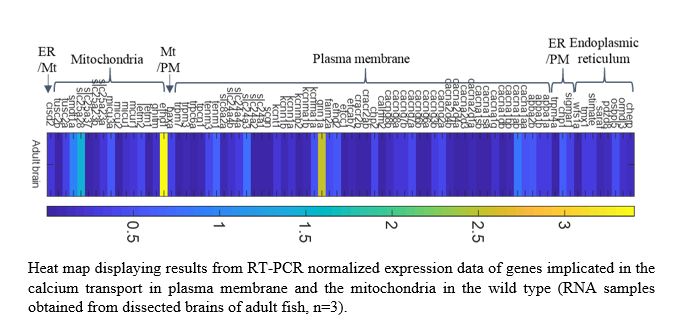
| gene symbol | location | human ortholog | expression data | |
| adult brain expression | larvae head expression | |||
| apba1a | PM | APBA1 | LOW | VERY LOW |
| apba1b | PM | APBA1 | LOW | VERY LOW |
| apba2b | PM | APBA2 | LOW | VERY LOW |
| apbb1 | C | APBB1 | VERY LOW | VERY LOW |
| apbb1ip | C | APBB1IP | Thisse et al., 2004 | |
| apbb2b | C | APBB2 | MEDIUM | LOW |
| apbb3 | C | APBB3 | VERY LOW | VERY LOW |
| atox1 | C | ATOX1 | MEDIUM | HIGH |
| atp2a3 | ER | ATP2A3 | Takeuchi et al., 2017 | |
| atp2b1a | PM | ATP2B1 | Liao et al., 2007; Thisse et al., 2004; Rauch et al., 2003; Thisse et al., 2001 | |
| atp2b2 | PM | ATP2B | Liao et al., 2007 | |
| atp2b3a | PM | ATP2B3 | Liao et al., 2007; Thisse et al., 2004 | |
| atp2b3b | PM | ATP2B3 | Liao et al., 2007 | |
| atp2b4 | PM | ATP2B4 | Liao et al., 2007 | |
| atp2c1 | G | ATP2C1 | Xia et al., 2017 | |
| atp6v0cb | PM | ATP6V0C | Chung et al., 2010; Hwang et al., 2009; Rauch et al., 2003 | |
| baxa | PM/Mt | BAX | LOW | LOW |
| cabp1a | PM | CABP1 | Thisse et al., 2004 | |
| cacna1aa | PM | CACNA1A | Thisse et al., 2005 | |
| LOW | VERY LOW | |||
| cacna1ab | PM | CACNA1A | MEDIUM | VERY LOW |
| cacna1ba | PM | CACNA1B | Jelen et al., 2007 | |
| cacna1bb | PM | CACNA1B | VERY LOW | VERY LOW |
| cacna1db | PM | CACNA1D | VERY LOW | VERY LOW |
| cacna1g | PM | CACNA1G | LOW | VERY LOW |
| cacna1sa | PM | CACNA1S | VERY LOW | VERY LOW |
| cacna1sb | PM | CACNA1S | VERY LOW | VERY LOW |
| cacna2d1a | PM | CACNA2D1 | VERY LOW | VERY LOW |
| cacna2d3 | PM | NA | VERY LOW | VERY LOW |
| cacna2d4a | PM | CACNA2D4 | VERY LOW | VERY LOW |
| cacna2d4b | PM | CACNA2D4 | VERY LOW | VERY LOW |
| cacnb2a | PM | CACNB2 | Ebert et al., 2008; Zhou et al., 2008 | |
| cacnb2b | PM | CACNB2 | Ebert et al., 2008; Zhou et al., 2008 | |
| cacnb3a | PM | CACNB3 | Zhou et al., 2008 | |
| cacnb4a | PM | CACNB4 | Ebert et al., 2008; Zhou et al., 2008 | |
| cacnb4b | PM | CACNB4 | Ebert et al., 2008; Zhou et al., 2008 | |
| cacng2a | PM | CACNG2 | Roy et al., 2016; Thisse et al., 2004 | |
| LOW | VERY LOW | |||
| cacng3b | PM | CACNG3 | LOW | VERY LOW |
| cacng6a | PM | CACNG6 | VERY LOW | VERY LOW |
| cacng6b | PM | CACNG6 | VERY LOW | VERY LOW |
| cacng7a | PM | CACNG7 | LOW | VERY LOW |
| cacng7b | PM | CACNG7 | LOW | VERY LOW |
| cacng8a | PM | CACNG8 | LOW | VERY LOW |
| cacng8b | PM | CACNG8 | VERY LOW | VERY LOW |
| calb2a | C | CALB2 | Bhoyar et al., 2017; Thisse et al., 2004; Rauch et al., 2003 | |
| calb2b | C | CALB2 | Bhoyar et al., 2017; Hortopan et al., 2010; Bae et al., 2009; Yokoi et al., 2009; Duggan et al., 2008; Hendricks et al., 2007; Rohrschneider et al., 2007 |
|
| calhm2 | PM | CALHM2 | VERY LOW | VERY LOW |
| calr | ER | CALR | Thisse et al., 2004 | |
| canx | ER | CANX | Hung et al., 2013 | |
| cgref1 | EM | CGREF1 | MEDIUM | MEDIUM |
| cherp | ER/C | CHERP | VERY LOW | VERY LOW |
| chp1 | PM/ER/N/C | CHP1 | MEDIUM | LOW |
| chp2 | PM/N | CHP2 | LOW | LOW |
| cisd2 | ER/Mt | CISD2 | LOW | LOW |
| cracr2ab | PM | CRACR2A | VERY LOW | VERY LOW |
| cracr2b | PM | CRACR2B | VERY LOW | VERY LOW |
| crebbpa | C | CREBBP | LOW | VERY LOW |
| crtc1a | C | CRTC1 | LOW | VERY LOW |
| crtc1b | C | CRTC1 | LOW | VERY LOW |
| crtc3 | C | CRTC3 | LOW | VERY LOW |
| efcab1 | C | EFCAB1 | VERY LOW | VERY LOW |
| efcab11 | C | EFCAB11 | VERY LOW | VERY LOW |
| efcab7 | PM | EFCAB7 | VERY LOW | VERY LOW |
| efcc1 | PM | EFCC1 | VERY LOW | VERY LOW |
| efhc2 | C | EFHC2 | VERY LOW | VERY LOW |
| efhd1 | Mt | EFHD1 | HIGH | MEDIUM |
| efhd2 | PM | EFHD2 | LOW | LOW |
| faim2a | PM | FAIM2 | LOW | VERY LOW |
| ghitm | Mt | GHITM | VERY LOW | VERY LOW |
| grin1a | PM | GRIN1 | Tzeng et al., 2007; Cox et al., 2005 | |
| HIGH | LOW | |||
| grin1b | PM | GRIN1 | Tzeng et al., 2007; Cox et al., 2005 | |
| grinab | PM | GRINA | Rojas-Rivera et al., 2012; Thisse et al., 2004 | |
| itpr3 | ER | ITPR3 | Petko et al., 2009 | |
| kcnma1a | PM | KCNMA1 | LOW | VERY LOW |
| kcnma1b | PM | KCNMA1 | VERY LOW | VERY LOW |
| kcnmb2 | PM | KCNMB2 | VERY LOW | VERY LOW |
| kcnn1a | PM | KCNN1 | VERY LOW | VERY LOW |
| kcnn1b | PM | KCNN1 | Cabo et al., 2013 | |
| VERY LOW | VERY LOW | |||
| kcnt1 | PM | KCNT1 | LOW | VERY LOW |
| letm1 | Mt | LETM1 | VERY LOW | VERY LOW |
| letm2 | Mt | LETM2 | LOW | LOW |
| mcu | Mt | MCU | Soman et al., 2017 | |
| mcur1 | Mt | MCUR1 | LOW | LOW |
| micu1 | Mt | MICU1 | VERY LOW | VERY LOW |
| micu2 | Mt | MICU2 | VERY LOW | VERY LOW |
| micu3a | Mt | MICU3 | LOW | LOW |
| ncs1a | PM | NCS1 | Petko et al., 2009; Blasiole et al., 2005; Thisse et al., 2004 | |
| ncs1b | PM | NCS1 | Blasiole et al., 2005 | |
| necab1 | C | NECAB1 | Kim et al., 2008; Thisse et al., 2004 | |
| necab2 | PM | NECAB2 | Kim et al., 2008; Thisse et al., 2004 | |
| ormdl3 | ER | ORMDL3 | LOW | VERY LOW |
| osbpl5 | C/ER | OSBPL5 | LOW | VERY LOW |
| pdzd8 | ER | PDZD8 | VERY LOW | VERY LOW |
| ppef1 | C | PPEF1 | VERY LOW | VERY LOW |
| ryr2a | ER | RYR2 | Wu et al., 2011 | |
| saraf | ER | SARAF | VERY LOW | MEDIUM |
| scgn | PMC | SCGN | VERY LOW | VERY LOW |
| sigmar1 | ER/PM/N/C | SIGMAR1 | LOW | LOW |
| slc24a1 | PM | SLC24A1 | VERY LOW | VERY LOW |
| slc24a2 | PM | SLC24A2 | VERY LOW | LOW |
| slc24a3 | PM | SLC24A3 | MEDIUM | MEDIUM |
| slc24a4a | PM | SLC24A4 | Thisse et al., 2004 | |
| LOW | LOW | |||
| slc24a4b | PM | SLC24A4 | LOW | LOW |
| slc25a23a | Mt | SLC25A23/SLC25A41 | LOW | LOW |
| slc25a23b | Mt | SLC25A23/SLC25A41 | VERY LOW | VERY LOW |
| slc25a37 | Mt | SLC25A37 | VERY LOW | VERY LOW |
| slc25a28 | Mt | SLC25A28 | HIGH | HIGH |
| slc8a1b | PM | SLC8A1 | Liao et al., 2007; Langenbacher et al., 2005 | |
| slc8a2a | PM | SLC8A2 | Liao et al., 2007 | |
| VERY LOW | VERY LOW | |||
| slc8a2b | PM | SLC8A2 | Liao et al., 2007 | |
| slc8a3 | PM | SLC8A3 | Liao et al., 2007 | |
| slc8a4a | PM | NA | On et al., 2009; Liao et al., 2007 | |
| slc8a4b | PM | NA | Liao et al., 2007 | |
| smdt1a | Mt | SMDT1 | MEDIUM | MEDIUM |
| stimate | ER | STIMATE | LOW | LOW |
| tenm1 | PM/N | TENM1 | MEDIUM | MEDIUM |
| tenm3 | PM | TENM3 | Hortopan et al., 2010; Kudoh et al., 2001; Thisse et al., 2001; Mieda et al., 1999 | |
| LOW | LOW | |||
| tmbim4 | G | TMBIM4 | HIGH | HIGH |
| tmx1 | ER | TMX1 | LOW | LOW |
| tpcn1 | PM | TPCN1 | VERY LOW | VERY LOW |
| tpcn2 | L | TPCN2 | VERY LOW | VERY LOW |
| trpa1a | PM | TRPA1 | Prober et al., 2008 | |
| trpa1b | PM | TRPA1 | Gau et al., 2017; Faucherre et al., 2013; Gau et al., 2013; Pan et al., 2012; Caron et al., 2008; Prober et al., 2008 |
|
| trpc3 | PM | TRPC3 | von Niederhäusern et al., 2013 | |
| trpc4apa | PM | TRPC4 | von Niederhäusern et al., 2013 | |
| trpc6a | PM | TRPC6 | VERY LOW | VERY LOW |
| trpc7b-2 | PM | TRPC7 | von Niederhäusern et al., 2013 | |
| trpm2 | PM | TRPM2 | Kastenhuber et al., 2013 | |
| trpm3 | PM | TRPM3 | Kastenhuber et al., 2013 | |
| VERY LOW | VERY LOW | |||
| trpm4a | PM/ER/G | TRPM4 | VERY LOW | VERY LOW |
| trpm6 | PM | TRPM6 | Arjona et al., 2013 | |
| trpm7 | PM | TRPM7 | VERY LOW | VERY LOW |
| trpv1 | PM | TRPV1 | Gau et al., 2017; Gau et al., 2013; Graham et al., 2013; Pan et al., 2012; Caron et al., 2008 | |
| trpv4 | PM | TRPV4 | Graham et al., 2013; Amato et al., 2012; Mangos et al., 2007 | |
| trpv6 | PM | TRPV6 | Shu et al., 2016; Graham et al., 2013; Pan et al., 2005 | |
| tusc2a | Mt | TUSC2 | MEDIUM | LOW |
| tusc2b | Mt | TUSC2 | VERY LOW | LOW |
| vdac1 | Mt | VDAC1 | Gebriel et al., 2014; Thisse et al., 2004 | |
| vdac3 | Mt | VDAC3 | Thisse et al., 2004 | |
| wfs1a | ER | WFS1 | VERY LOW | VERY LOW |
PM - plasma membrane, C - cytoplasm, ER - endoplasmic reticullum, G - Golgi apparatus, Mt - mitochondria, N - nuclei, L - lysosome