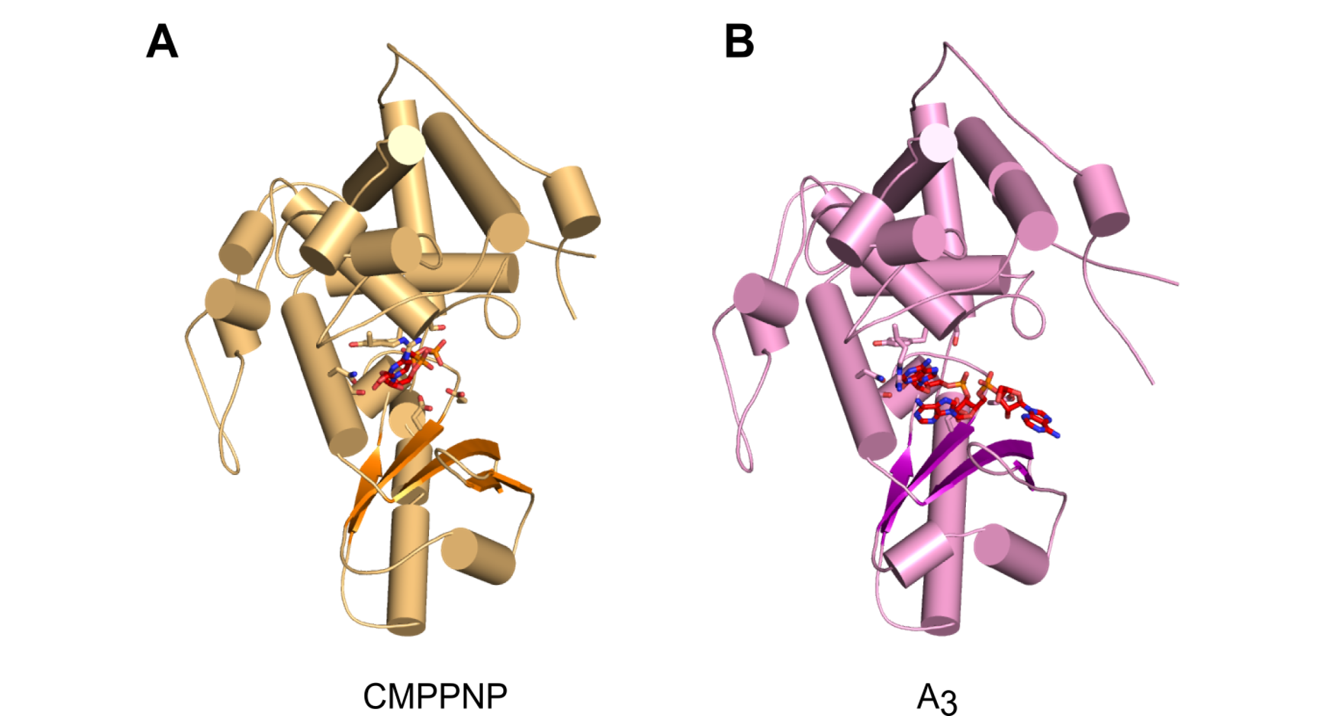Prof. Andrzej Dziembowski participated in Hypothalamus Gordon Research Conference. Unraveling the Complexity of the Hypothalamus from Single Molecules to Intricate Behaviors; July 24-29, 2022, Ventura, CA, United States with a poster titled "mRNAs of hypothalamic neuropeptades are polyadenylated in the cytoplasm"

We are pleased to inform that in 2022 we established cooperation with the BioCentre of Science Education (BioCEN) in the frame of the GRIEG projects. Our cooperation may cover publicly available laboratory workshops, trainings, presentations and lectures for adolescents, biology teachers, schools, companies and for all who are interested.
BioCentre of Science Education was established in 2002. It is a unique initiative popularizing science. It involves innovative methods, reaches a wide audience, it has its own laboratory in Warsaw and co-organizes events all over Poland for schools, companies, science festivals and other institutions, aiming at reaching teachers and students from smaller towns and villages.

Miło nam poinformować, że w 2022 roku nawiązaliśmy współpracę z BioCentrum Edukacji Naukowej (BioCEN) w ramach projektów GRIEG. Nasza współpraca może obejmować ogólnodostępne warsztaty laboratoryjne, szkolenia, prezentacje i wykłady dla młodzieży, nauczycieli biologii, szkół, firm i dla wszystkich zainteresowanych.
BioCentrum Edukacji Naukowej powstało w 2002 roku. Jest unikalną inicjatywą popularyzującą naukę. Stosuje innowacyjne metody, dociera do szerokiego grona odbiorców, posiada własne laboratorium w Warszawie oraz współorganizuje wydarzenia w całej Polsce dla szkół, firm, festiwali nauki i innych instytucji, mając na celu dotarcie do nauczycieli i uczniów z mniejszych miast i wsi.
W dniach 18-20 października 2022 roku członkowie Laboratorium Biologii RNA - Grupa ERA Chairs pod kierownictwem prof. Andrzeja Dziembowskiego (koordynatora projektu) spotkali się z partnerami: Centrum Nowych Technologii, Uniwersytet Warszawski (Polska) oraz Uniwersytet w Bergen (Norwegia).
Podczas spotkania omówiono postępy i przyszłe kierunki badań finansowanych z grantu GRIEG.
 |
|
On October 18-20, 2022 members of the Laboratory of RNA Biology - ERA Chairs Group headed by Prof. Andrzej Dziembowski (coordinator of the project) met with partners: the Centre of New Technologies, University of Warsaw, Poland and University of Bergen, Norway.
During the meeting progress and future directions of research funded by the GRIEG grant was discussed.
 |
 |
Drug development projects
We cooperated with pharmaceutical companies in multiple drug development projects by performing structural studies of complexes of target proteins with inhibitors. Until the end of 2019 these projects were performed by a specialized laboratory – IIMCB Structural Biology Center with Marcin Nowotny as a CSO.
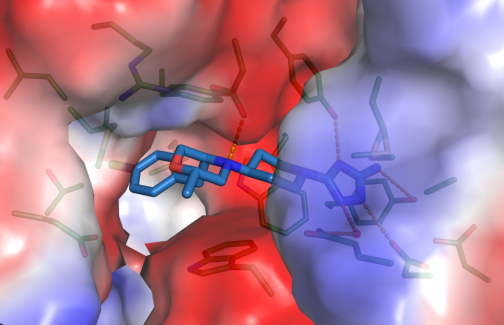
A published example of such cooperation concerns OATD-01 compound developed by OncoArendi Therapeutics company. OATD-01 is an inhibitor chitotriosidase/acidic mammalian chitinase which can be used in the treatment of idiopathic pulmonary fibrosis. The compound recently completed phase I clinical studies. We contributed the structural characterization of the complex between chitinase and the inhibitor.
Robert Koralewski, Barbara Dymek, Marzena Mazur, Piotr Sklepkiewicz, Sylwia Olejniczak, Wojciech Czestkowski, Krzysztof Matyszewski, Gleb Andryianau, Piotr Niedziejko, Michal Kowalski, Mariusz Gruza, Bartłomiej Borek, Karol Jedrzejczak, Agnieszka Bartoszewicz, Elżbieta Pluta, Aleksandra Rymaszewska, Magdalena Kania, Tomasz Rejczak, Sylwia Piasecka, Michal Mlacki, Marcin Mazurkiewicz, Michał Piotrowicz, Magdalena Salamon, Agnieszka Zagozdzon, Agnieszka Napiorkowska-Gromadzka, Aneta Bartlomiejczak, Witold Mozga, Paweł Dobrzański, Karolina Dzwonek, Jakub Golab, Marcin Nowotny, Jacek Olczak, and Adam Golebiowski, Discovery of OATD-01, a First-in-Class Chitinase Inhibitor as Potential New Therapeutics for Idiopathic Pulmonary Fibrosis. J Med Chem 2020 Dec 24;63(24):15527-15540. Published October 20, 2020, https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c01179#
Close-up view of active site of hCHIT1 (represented as surface colored according to electrostatic potential) in complex with OATD-01 (blue sticks).
Nucleic acid-based therapeutics
Antisense technology is an emerging therapeutic approach for inhibiting gene expression through recognition and RNase H1-dependent degradation of cellular mRNAs. In this technology, short, synthetic, single-stranded oligonucleotides (ASOs) are designed that are complementary to the target mRNA.
ASOs are frequently chemically modified for enhancing the therapeutic properties of these molecules. The most widely used modifications include phosphorothioate (PS) backbone and modifications of the sugar moieties at 2’ site. The molecular mechanism of toxicity of chemically modified antisense oligonucleotides are not fully understood.
Hyjek-Składanowska M, Vickers T, Napiórkowska A, Anderson BA, Tanowitz M, Crooke ST, Liang X, Seth PP&, Nowotny M&. Origins of the increased affinity of phosphorothioate-modified therapeutic nucleic acids for proteins. J. Am. Chem. Soc., 2020, 142 (16): 7456-7468;
& - corresponding authors
-
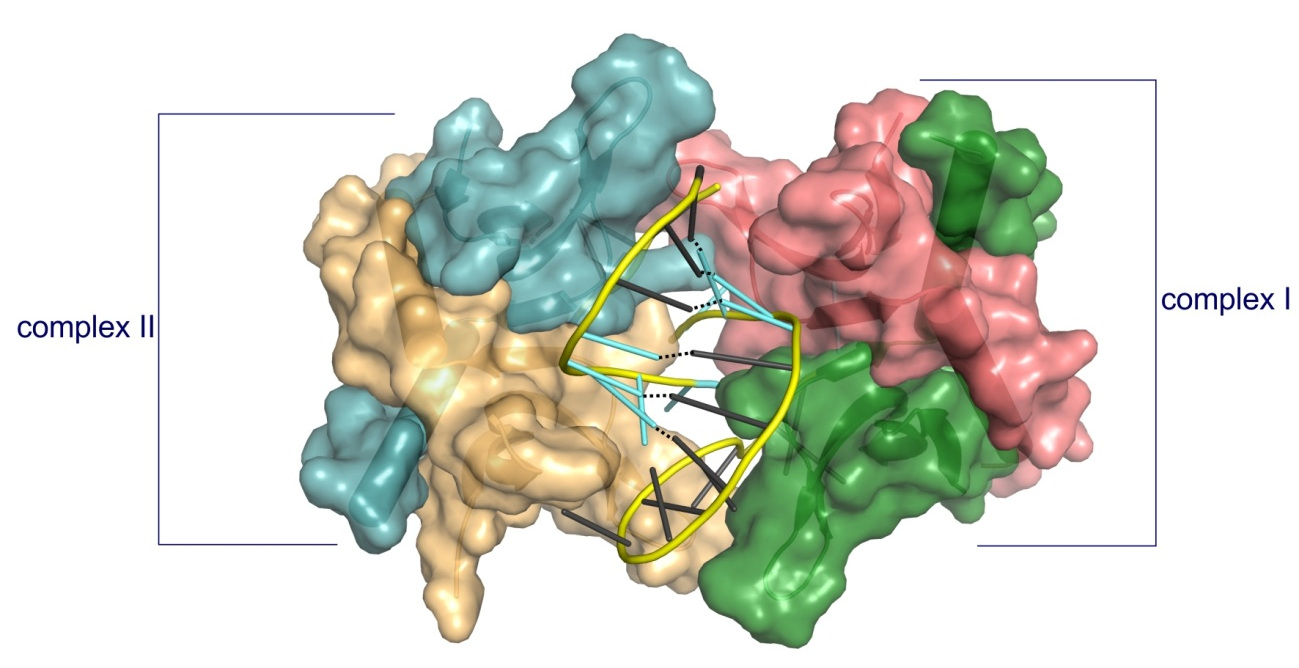
The first crystal structure of a complex between a protein and fully PS nucleic acid (DNA-binding domain of a model ASO-binding protein PC4 in complex with full PS 2′-OMe DNA gapmer ASO).
-
The structure reveals a possible mechanism of ASO-induced toxic protein aggregation: ASO is bound in hairpin-like conformation and its exposed gapmer part promotes the formation of a dimer of dimers of PC4 through base pairing.
-
The protein interacts with the PS-nucleic acid through a network of electrostatic and hydrophobic interactions. Importantly, the backbone of the PS ASO is able to form new and more extensive hydrophobic interactions than a natural phosphodiester backbone which provides insights into the origins for the enhanced affinity of PS for proteins.
The studies of PC4-ASO complex have been performed in cooperation with Ionis Pharmaceuticals (Carlsbad, California, USA).
Structure of PC4 in complex with PS 2′-OMe DNA gapmer ASO. The content of the asymmetric unit of the PTEN ASO complex crystal. PC4 protomers are color-coded and shown in surface representation, DNA is shown in cartoon (PS backbone is shown in yellow, 2′-OMe PS nucleotides are shown in aquamarine, DNA gapmer nucleotides are shown in dark gray). The base pairing between the nucleotides is shown as black dotted lines.
Hyjek-Składanowska M, Anderson BA, Mykhaylyk V, Orr C, Wagner A, Poznański JT, Skowronek K, Seth PP, Nowotny M. Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding. Nucleic Acids Research, 2022;, gkac774, https://doi.org/10.1093/nar/gkac774
-
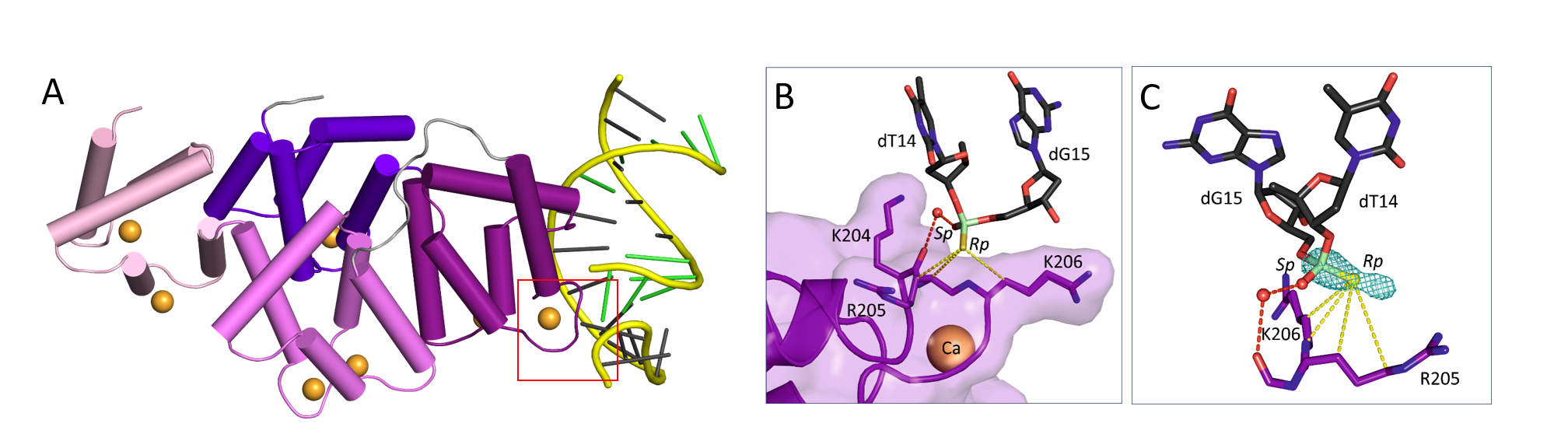
The first crystal structure of a complex between a PS oligonucleotide and non-specific nucleic acids binding protein (core domain of annexin A2).
-
Interactions between the sulfur atom of the PS linkage and the protein surface are mainly of hydrophobic character - the hydrophobic nature of sulfur contributes to the association of PS ASOs with proteins.
-
Measurements of anomalous diffraction of sulfur confirmed that van der Waals contacts between the sulfur atom and hydrophobic parts of arginine and lysine side chains drive the enhanced interaction of PS ASOs with proteins.
-
Stereoisomer preference at given phosphorothioate in the DNA oligonucleotide is determined by the environment around the PS linkage coming from the protein and other adjacent structural features such as 5-Me groups on cytosine nucleobases.
The studies of AnxA2-ASO complex have been performed in cooperation with Ionis Pharmaceuticals (Carlsbad, California, USA) and Diamond synchrotron (Didcot, Oxfordshire, UK).
(A) Crystal structure of AnxA2 in complex with PS ASO duplex. Calcium-binding annexing domain I-IV are coloured light pink, violet, purple, and purple blue, respectively. Calcium ions are shown as orange spheres. DNA is shown in cartoon (PS backbone is shown in yellow, 2’-MOE PS nucleotides are shown in green, DNA gapmer nucleotides are shown in dark gray). (B) Close-up view on the phosphorothioate-binding surface in AnxA2. Polar interactions are shown as red dotted lines. Van der Waals interactions are shown as yellow dotted lines. (C) Difference Fourier anomalous map calculated based on long-wavelength X-ray diffraction data (λ = 2.7552 Ȧ), shown as teal mesh. The preferred occupancy of Rp PS stereoisomer is facilitated by the hydrophobic interactions with surrounding amino acids.
RNA processing
Human cap 2’-OH methyltransferase
mRNA contains a cap structure on its 5' terminus. In higher eukaryotes the first and the second ribonucleotide of the body of the mRNA are methylated on the 2' oxygen. This is thought to serve as a tag to distinguish self mRNA from the mRNA of invading viruses. To circumvent the mechanism, some viruses such as West Nile and yellow fever viruses encode their own 2'-OH methyltransferases.
Śmietanski M*, Werner M*, Purta E, Kamińska KH, Stepiński J, Darżynkiewicz E, Nowotny M&, Bujnicki JM&. Structural analysis of human 2’-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun., 2014 Jan 9;5:3004; * - equally contributing, & - corresponding authors
-
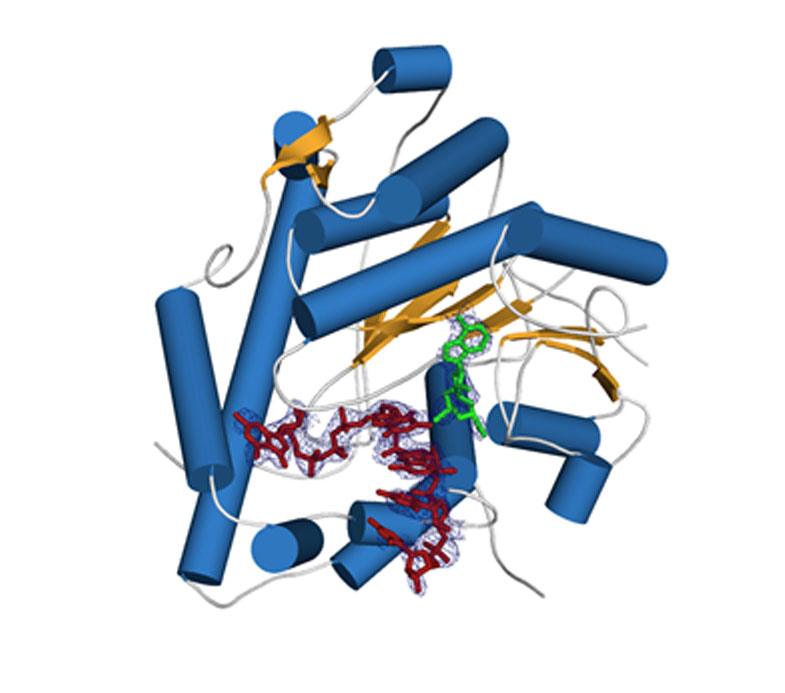
The first crystal structure of a cellular 2'-OH cap methyltransferase.
-
The mode of cap recognition markedly different from viral counterparts providing hints for antiviral drug development.
The studies of cap 2'-OH methyltransferase have been performed in collaboration with Prof. Janusz M.Bujnicki (IIMCB).
Crystal structure of human cap 2'-OH cap methyl transferase 1 (hMTR1). A fragment of capped mRNA is shown in red and the S-adenosylmethionine (methyl group donor) in green.
Mitochondrial exoribonuclease complex mtEXO
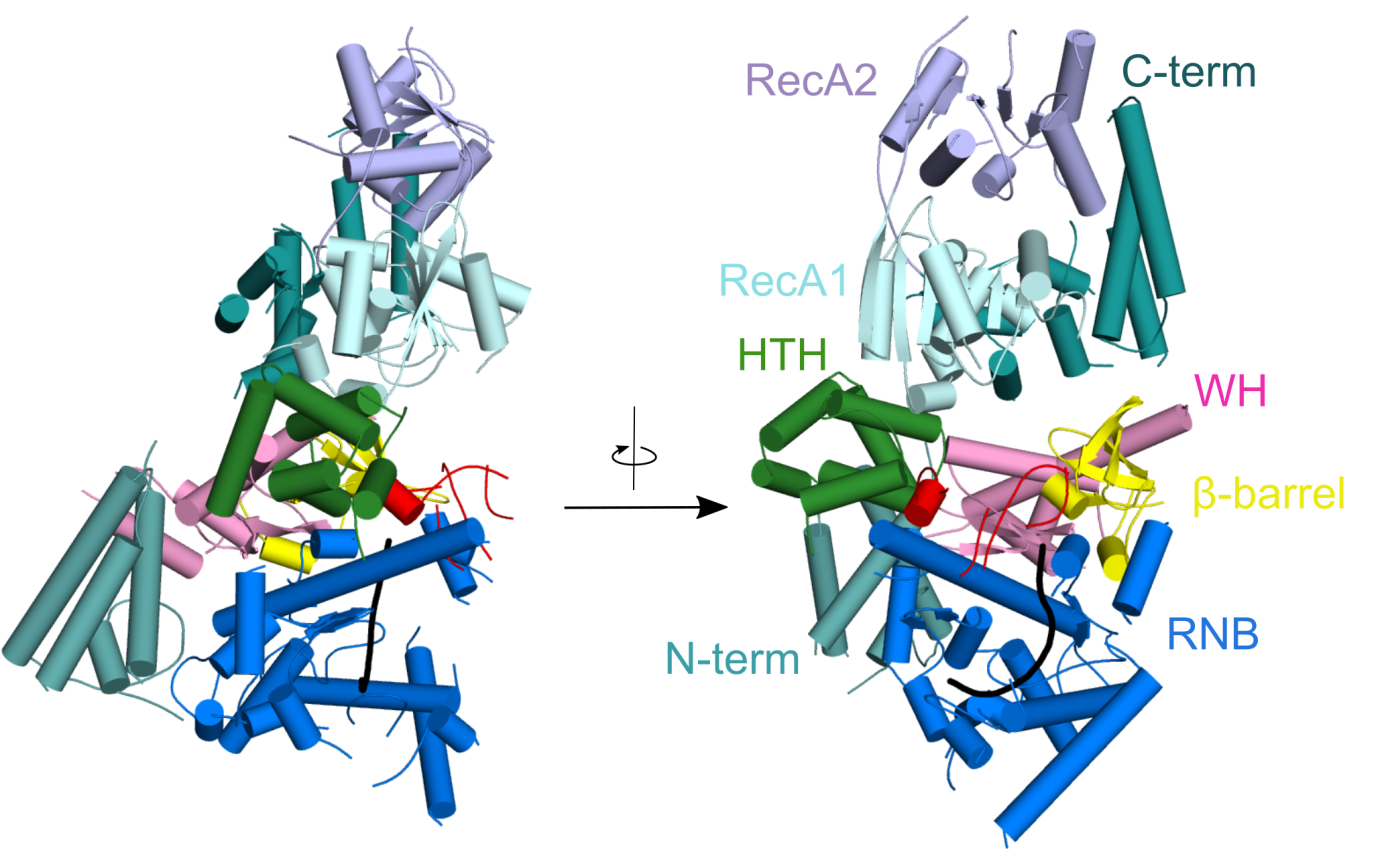
RNA degradation pathways play crucial roles in processing of various types of RNA, regulation of gene expression, and efficient removal of defective RNAs. The main executor of RNA turnover and surveillance activity in yeast mitochondria is the mtEXO complex, composed of Dss1 3ʹ-to-5ʹ exoribonuclease and Suv3 helicase.
Rażew M, Warkocki Z, Taube M, Kolondra A, Czarnocki-Cieciura M, Nowak E, Łabędzka-Dmoch K, Kawińska A, Piątkowski J, Golik P, Kozak M, Dziembowski A, Nowotny M. Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components. Nat Commun., 2018 Jan 8;9(1):97.
-
Crystal structure of Dss1 exoribonuclease from Candida glabrata reveals it is a unique member of the RNase II family with specialized domains responsible for interactions with Suv3 helicase.
-
Crystal structure of the mtEXO complex reveals the arrangement of both subunits in which the helicase motor feeds the 3' end of the RNA into the catalytic channel of Dss1 for its efficient degradation.
-
Co-operation of both helicase and nuclease activities within the complex is particularly important for degeneration of structured RNAs which cannot be handled by Dss1 on its own and for which the unwinding activity of Suv3 is required.
Crystal structure of Candida glabrata mtEXO complex shows the arrangement of the Suv3 helicase on top of Dss1 exoribonucleases and its accessory domains decorating the catalytic RNB domain (shown in blue) with the RNA molecule trapped inside (shown in black).
CutA terminal ribonucleotide transferase
Template-independent terminal ribonucleotide transferases (TENTs) catalyze the addition of nucleotide monophosphates to the 3′-end of RNA molecules regulating their fate. A subgroup TENTs are 3′ CUCU-tagging enzymes, such as CutA in Aspergillus nidulans. CutA preferentially incorporates cytosines, processively polymerizes only adenosines and does not incorporate or extend guanosines.
Malik D, Kobyłecki K, Krawczyk P, Poznański J, Jakielaszek A, Napiórkowska A, Dziembowski A, Tomecki R&, Nowotny M&, Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine, Nucleic Acids Res., 2020, 48(16):9387-9405; & - corresponding authors
-
The first structural characterization of a TENT adding C/U tails - structures solved for CutA in complex with incoming CTP analog and RNA with three adenosines.
-
The binding of GTP or a primer with a terminal guanosine is predicted to lead to clashes between NH2 of the guanine and the protein, explaining why CutA is unable to use these ligands as substrates.
-
Processive adenosine incorporation likely results from tighter binding of the primer with 3′-terminal adenosine and efficient stacking between adenosine bases of the primer and incoming ATP.
-
A dynamic process of NTP recognition proposed.
Overall structures of CutA complexes. (A) Structure of CutA–CMPCPP complex. The protein is shown as an orange cartoon, with β-strands in a darker shade of orange. Incoming nucleotide is shown as red sticks and active site residues are shown as orange sticks. (B) Structure of CutA–A3 complex. The protein is shown as pink cartoon, with β-strands in purple. The RNA is shown as red sticks and active site residues are shown as pink sticks.