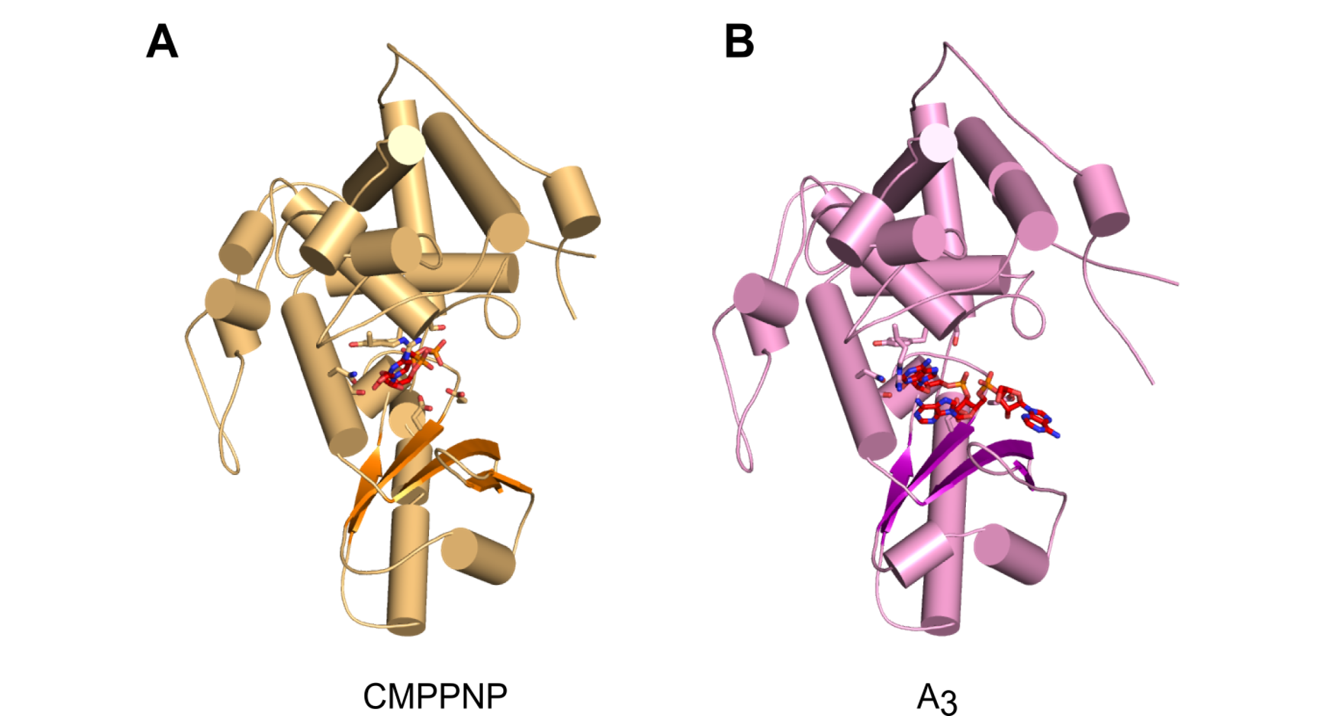RNA processing
Human cap 2’-OH methyltransferase
mRNA contains a cap structure on its 5' terminus. In higher eukaryotes the first and the second ribonucleotide of the body of the mRNA are methylated on the 2' oxygen. This is thought to serve as a tag to distinguish self mRNA from the mRNA of invading viruses. To circumvent the mechanism, some viruses such as West Nile and yellow fever viruses encode their own 2'-OH methyltransferases.
Śmietanski M*, Werner M*, Purta E, Kamińska KH, Stepiński J, Darżynkiewicz E, Nowotny M&, Bujnicki JM&. Structural analysis of human 2’-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun., 2014 Jan 9;5:3004; * - equally contributing, & - corresponding authors
-
The first crystal structure of a cellular 2'-OH cap methyltransferase.
-
The mode of cap recognition markedly different from viral counterparts providing hints for antiviral drug development.
The studies of cap 2'-OH methyltransferase have been performed in collaboration with Prof. Janusz M.Bujnicki (IIMCB).
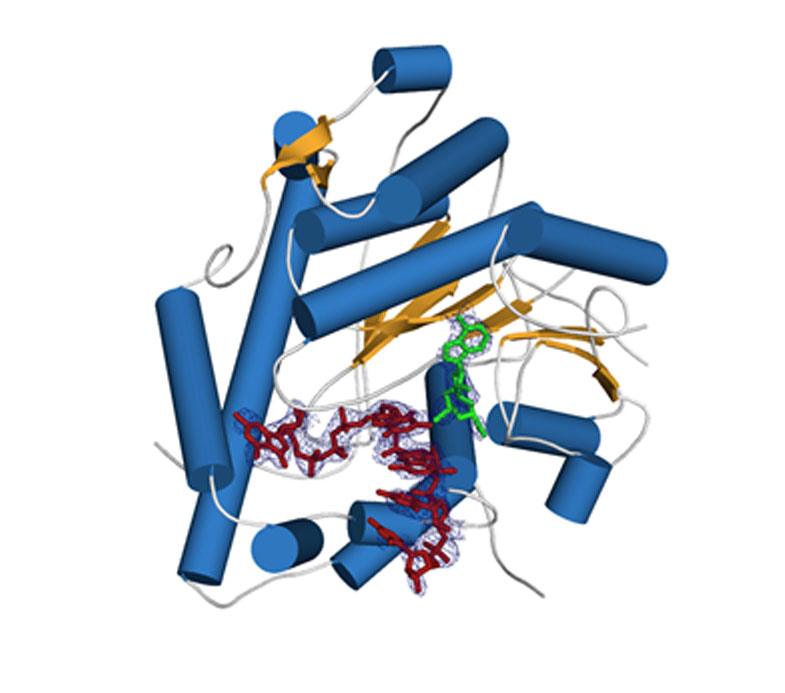
Crystal structure of human cap 2'-OH cap methyl transferase 1 (hMTR1). A fragment of capped mRNA is shown in red and the S-adenosylmethionine (methyl group donor) in green.
Mitochondrial exoribonuclease complex mtEXO
RNA degradation pathways play crucial roles in processing of various types of RNA, regulation of gene expression, and efficient removal of defective RNAs. The main executor of RNA turnover and surveillance activity in yeast mitochondria is the mtEXO complex, composed of Dss1 3ʹ-to-5ʹ exoribonuclease and Suv3 helicase.
Rażew M, Warkocki Z, Taube M, Kolondra A, Czarnocki-Cieciura M, Nowak E, Łabędzka-Dmoch K, Kawińska A, Piątkowski J, Golik P, Kozak M, Dziembowski A, Nowotny M. Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components. Nat Commun., 2018 Jan 8;9(1):97.
-
Crystal structure of Dss1 exoribonuclease from Candida glabrata reveals it is a unique member of the RNase II family with specialized domains responsible for interactions with Suv3 helicase.
-
Crystal structure of the mtEXO complex reveals the arrangement of both subunits in which the helicase motor feeds the 3' end of the RNA into the catalytic channel of Dss1 for its efficient degradation.
-
Co-operation of both helicase and nuclease activities within the complex is particularly important for degeneration of structured RNAs which cannot be handled by Dss1 on its own and for which the unwinding activity of Suv3 is required.
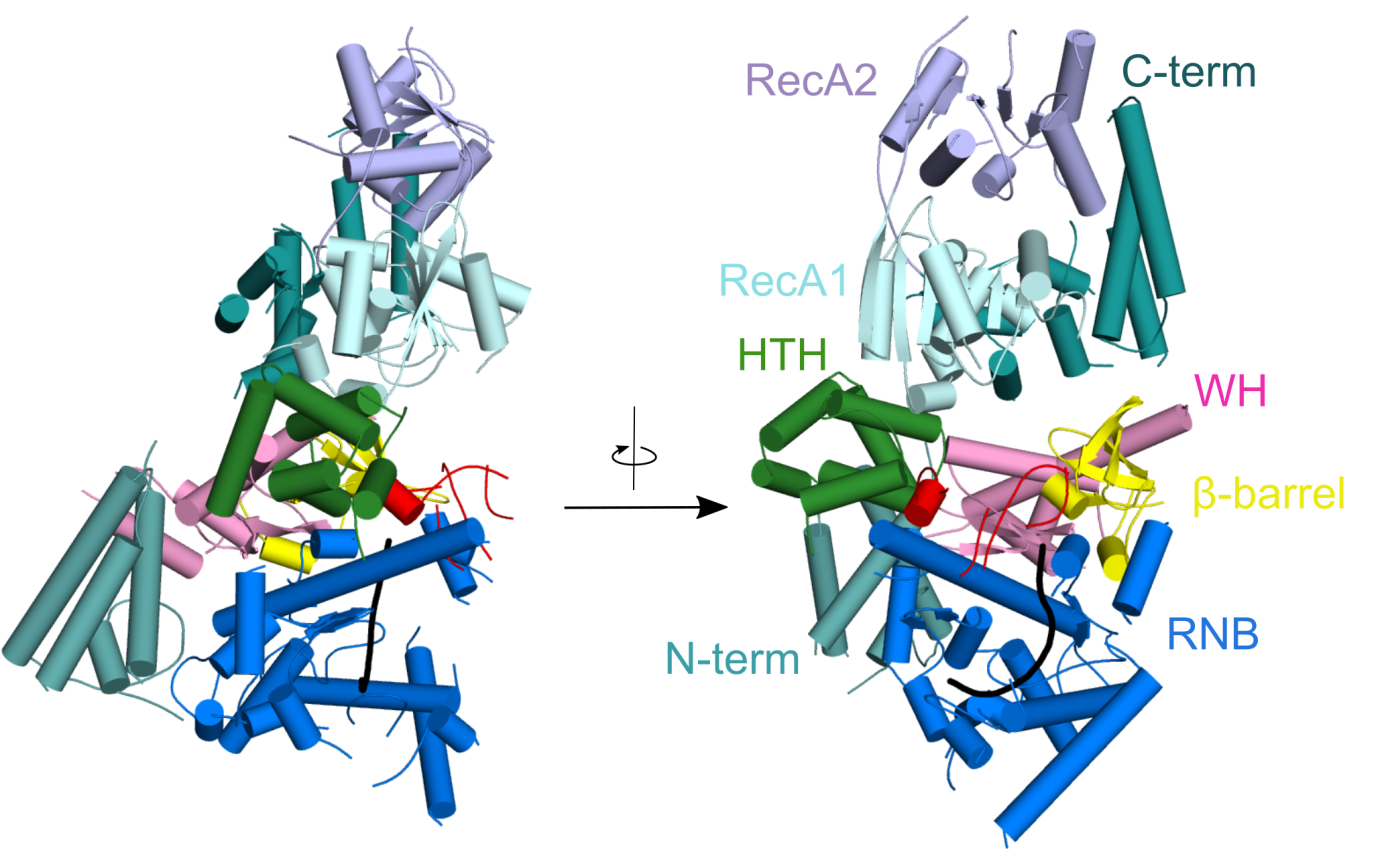
Crystal structure of Candida glabrata mtEXO complex shows the arrangement of the Suv3 helicase on top of Dss1 exoribonucleases and its accessory domains decorating the catalytic RNB domain (shown in blue) with the RNA molecule trapped inside (shown in black).
CutA terminal ribonucleotide transferase
Template-independent terminal ribonucleotide transferases (TENTs) catalyze the addition of nucleotide monophosphates to the 3′-end of RNA molecules regulating their fate. A subgroup TENTs are 3′ CUCU-tagging enzymes, such as CutA in Aspergillus nidulans. CutA preferentially incorporates cytosines, processively polymerizes only adenosines and does not incorporate or extend guanosines.
Malik D, Kobyłecki K, Krawczyk P, Poznański J, Jakielaszek A, Napiórkowska A, Dziembowski A, Tomecki R&, Nowotny M&, Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine, Nucleic Acids Res., 2020, 48(16):9387-9405; & - corresponding authors
-
The first structural characterization of a TENT adding C/U tails - structures solved for CutA in complex with incoming CTP analog and RNA with three adenosines.
-
The binding of GTP or a primer with a terminal guanosine is predicted to lead to clashes between NH2 of the guanine and the protein, explaining why CutA is unable to use these ligands as substrates.
-
Processive adenosine incorporation likely results from tighter binding of the primer with 3′-terminal adenosine and efficient stacking between adenosine bases of the primer and incoming ATP.
-
A dynamic process of NTP recognition proposed.
Overall structures of CutA complexes. (A) Structure of CutA–CMPCPP complex. The protein is shown as an orange cartoon, with β-strands in a darker shade of orange. Incoming nucleotide is shown as red sticks and active site residues are shown as orange sticks. (B) Structure of CutA–A3 complex. The protein is shown as pink cartoon, with β-strands in purple. The RNA is shown as red sticks and active site residues are shown as pink sticks.
tRNA ligases
Some tRNA transcripts contain introns. After their excision the exons are ligated by RNA ligases RtcB and RTCB in archaea and metazoa, respectively. The metazoan RTCB forms a stable complex with four additional subunits: DDX1, FAM98B, CGI99, and ASHWIN.
Chamera S, Zajko W, Czarnocki-Cieciura M, Jaciuk M, Koziej Ł, Nowak J, Wycisk K, Sroka M, Chramiec-Głąbik A, Śmietański M, Gołębiowski F, Warmiński M, Jemielity J, Glatt S, Nowotny M. Structural and biochemical characterization of the 3'-5' tRNA splicing ligases. J. Biol. Chem.; 2025; 301(5):108506
- Thiol-based chemical cross-linking revealed the involvement of specific RtcB residues in RNA binding.
- 3.3 Å cryo-EM structure of the five-subunit Danio rerio tRNA ligase complex.
- FAM98B and CGI99 form a highly intertwined heterodimer with a helical bundle at the center of the complex that binds the C-terminus of DDX1 and the catalytic subunit, RTCB.
- The DDX1 helicase module is mobile and can modulate RTCB activity.
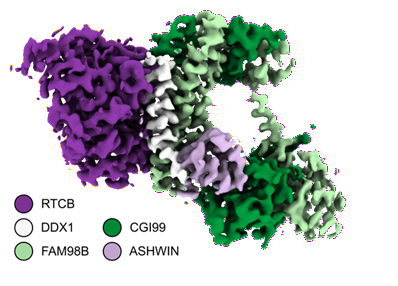
Cryo-EM structure of the five-subunit D. rerio tRNA ligase complex.
enDR3 – a tool for mapping RNA/DNA hybrids
R-loops are nucleic acid structures consisting of an RNA/DNA hybrid and a displaced single-stranded DNA that arise during transcription. Impaired processing of R-loops has been linked to genome instability, a hallmark of various severe human diseases. Despite their biological importance, the mechanisms governing R-loop formation and regulation remain poorly understood and new tools are needed to their precise localization in the genome.
Marta Jedynak-Slyvka*, Zuzanna Kaczmarska*, Damian Graczyk, Aneta Jurkiewicz, Małgorzata Figiel, Lukasz S Borowski, Roman J Szczesny&, Marcin Nowotny& Genome-wide in vivo and ex vivo mapping of R-loops using engineered N-terminal hybrid-binding domain of RNase H3 (enDR3), Nucleic Acids Res., 2025: 53(15); * - equally contributing, & - corresponding authors
-
enDR3 is a novel tool engineered to capture and sequence RNA/DNA hybrids with high precision for accurate R-loop localization.
-
It consists of an engineered tandem fusion of a modified N-terminal hybrid-binding domain derived from bacterial RNase H3.
-
enDR3 has been successfully utilized for genome-wide R-loop profiling through DNA/RNA immunoprecipitation coupled with complementary DNA conversion and chromatin immunoprecipitation techniques.
-
The tool enables robust detection of R-loops both ex vivo and in vivo, offering a versatile platform for advancing research in R-loop biology.
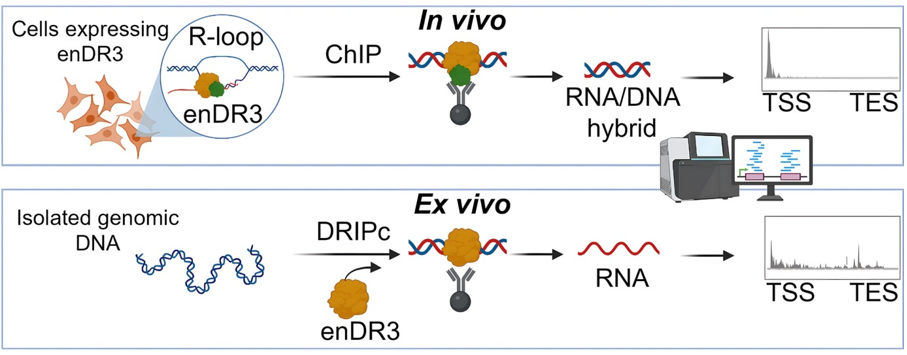
Schematic of the use of enDR3 in mapping RNA/DNA hybrids (R-loops) ex vivo and in vivo