W dniu 19 września 2022 Prof. A.Dziembowski wystąpił z prezentacją naukową w NIH RNA Biology Laboratory. Wystąpienie było zatytułowane „Ribonucleases and poly(A) polymerases in cancer and other diseases".
Prof. Andrzej Dziembowski participated in NIH RNA Biology Laboratory on September 19, 2022 with a presentation titled "Ribonucleases and poly(A) polymerases in cancer and other diseases".
W dniach 24-29 lipca 2022 Prof. Andrzej Dziembowski uczestniczył w konferencji pt. Hypothalamus Gordon Research, z wystąpieniem zatytułowanym „Unraveling the Complexity of the Hypothalamus from Single Molecules to Intricate Behaviors. Podczas wydarzenia, prof. Dziembowski zaprezentował poster zatytułowany „mRNAs of hypothalamic neuropeptades are polyadenylated in the cytoplasm”. Konferencja odbyła się w Ventura, w Stanach Zjednoczonych.
Prof. Andrzej Dziembowski participated in Hypothalamus Gordon Research Conference. Unraveling the Complexity of the Hypothalamus from Single Molecules to Intricate Behaviors; July 24-29, 2022, Ventura, CA, United States with a poster titled "mRNAs of hypothalamic neuropeptades are polyadenylated in the cytoplasm"

We are pleased to inform that in 2022 we established cooperation with the BioCentre of Science Education (BioCEN) in the frame of the GRIEG projects. Our cooperation may cover publicly available laboratory workshops, trainings, presentations and lectures for adolescents, biology teachers, schools, companies and for all who are interested.
BioCentre of Science Education was established in 2002. It is a unique initiative popularizing science. It involves innovative methods, reaches a wide audience, it has its own laboratory in Warsaw and co-organizes events all over Poland for schools, companies, science festivals and other institutions, aiming at reaching teachers and students from smaller towns and villages.

Miło nam poinformować, że w 2022 roku nawiązaliśmy współpracę z BioCentrum Edukacji Naukowej (BioCEN) w ramach projektów GRIEG. Nasza współpraca może obejmować ogólnodostępne warsztaty laboratoryjne, szkolenia, prezentacje i wykłady dla młodzieży, nauczycieli biologii, szkół, firm i dla wszystkich zainteresowanych.
BioCentrum Edukacji Naukowej powstało w 2002 roku. Jest unikalną inicjatywą popularyzującą naukę. Stosuje innowacyjne metody, dociera do szerokiego grona odbiorców, posiada własne laboratorium w Warszawie oraz współorganizuje wydarzenia w całej Polsce dla szkół, firm, festiwali nauki i innych instytucji, mając na celu dotarcie do nauczycieli i uczniów z mniejszych miast i wsi.
W dniach 18-20 października 2022 roku członkowie Laboratorium Biologii RNA - Grupa ERA Chairs pod kierownictwem prof. Andrzeja Dziembowskiego (koordynatora projektu) spotkali się z partnerami: Centrum Nowych Technologii, Uniwersytet Warszawski (Polska) oraz Uniwersytet w Bergen (Norwegia).
Podczas spotkania omówiono postępy i przyszłe kierunki badań finansowanych z grantu GRIEG.
 |
|
On October 18-20, 2022 members of the Laboratory of RNA Biology - ERA Chairs Group headed by Prof. Andrzej Dziembowski (coordinator of the project) met with partners: the Centre of New Technologies, University of Warsaw, Poland and University of Bergen, Norway.
During the meeting progress and future directions of research funded by the GRIEG grant was discussed.
 |
 |
Drug development projects
We cooperated with pharmaceutical companies in multiple drug development projects by performing structural studies of complexes of target proteins with inhibitors. Until the end of 2019 these projects were performed by a specialized laboratory – IIMCB Structural Biology Center with Marcin Nowotny as a CSO.
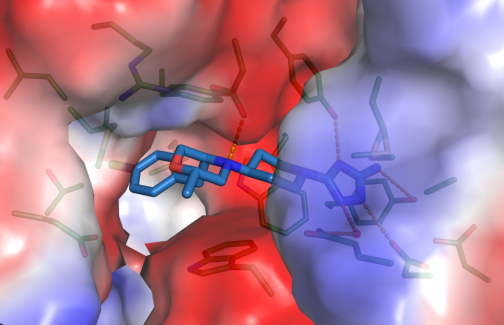
A published example of such cooperation concerns OATD-01 compound developed by OncoArendi Therapeutics company. OATD-01 is an inhibitor chitotriosidase/acidic mammalian chitinase which can be used in the treatment of idiopathic pulmonary fibrosis. The compound recently completed phase I clinical studies. We contributed the structural characterization of the complex between chitinase and the inhibitor.
Robert Koralewski, Barbara Dymek, Marzena Mazur, Piotr Sklepkiewicz, Sylwia Olejniczak, Wojciech Czestkowski, Krzysztof Matyszewski, Gleb Andryianau, Piotr Niedziejko, Michal Kowalski, Mariusz Gruza, Bartłomiej Borek, Karol Jedrzejczak, Agnieszka Bartoszewicz, Elżbieta Pluta, Aleksandra Rymaszewska, Magdalena Kania, Tomasz Rejczak, Sylwia Piasecka, Michal Mlacki, Marcin Mazurkiewicz, Michał Piotrowicz, Magdalena Salamon, Agnieszka Zagozdzon, Agnieszka Napiorkowska-Gromadzka, Aneta Bartlomiejczak, Witold Mozga, Paweł Dobrzański, Karolina Dzwonek, Jakub Golab, Marcin Nowotny, Jacek Olczak, and Adam Golebiowski, Discovery of OATD-01, a First-in-Class Chitinase Inhibitor as Potential New Therapeutics for Idiopathic Pulmonary Fibrosis. J Med Chem 2020 Dec 24;63(24):15527-15540. Published October 20, 2020, https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c01179#
Close-up view of active site of hCHIT1 (represented as surface colored according to electrostatic potential) in complex with OATD-01 (blue sticks).