Reverse transcriptases
XMRV reverse transcriptase
Reverse transcriptases use two enzymatic activities – DNA polymerase and RNase H to catalyze the conversion of single-stranded RNA to double-stranded DNA, a process essential for proliferation of retroviruses such as HIV and retrotransposons. Retroviral RTs are divided into two classes – dimeric (i. e. HIV) or monomeric (i. e. gammaretroviral enzyme from mouse Moloney leukemia virus and closely related XMRV).
Nowak E, Potrzebowski W, Konarev PV, Rausch JW, Bona MK, Svergun DI, Bujnicki JM, Le Grice SF, Nowotny M. Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid. Nucleic Acids Res., 2013 Apr 1;41(6):3874-87
-
The first crystal structure of a monomeric RTs in complex with RNA/DNA hybrid visualizing the polymerase-connection fragment of the enzyme.
-
Full-length protein modelled based on small-angle X-ray scattering data.
-
SAXS data demonstrated that the RNase H domain is mobile and only occasionally interacts with the substrate to cleave RNA. This is the mechanism to fine tune RNase H activity.
-
This mechanism of RNase H fine-tuning is different from dimeric RTs which use substrate deformations for that purpose.
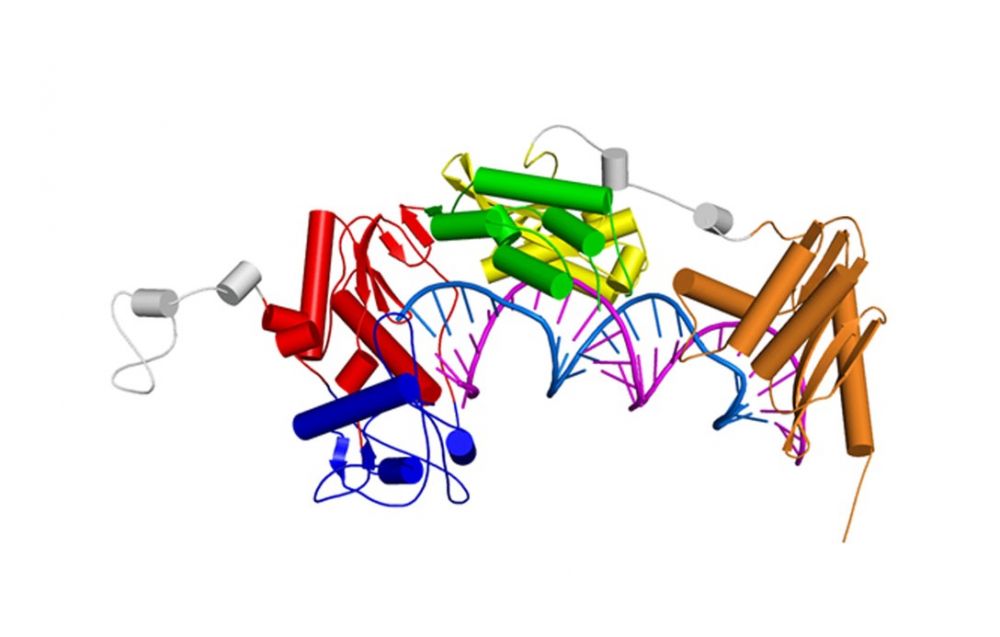
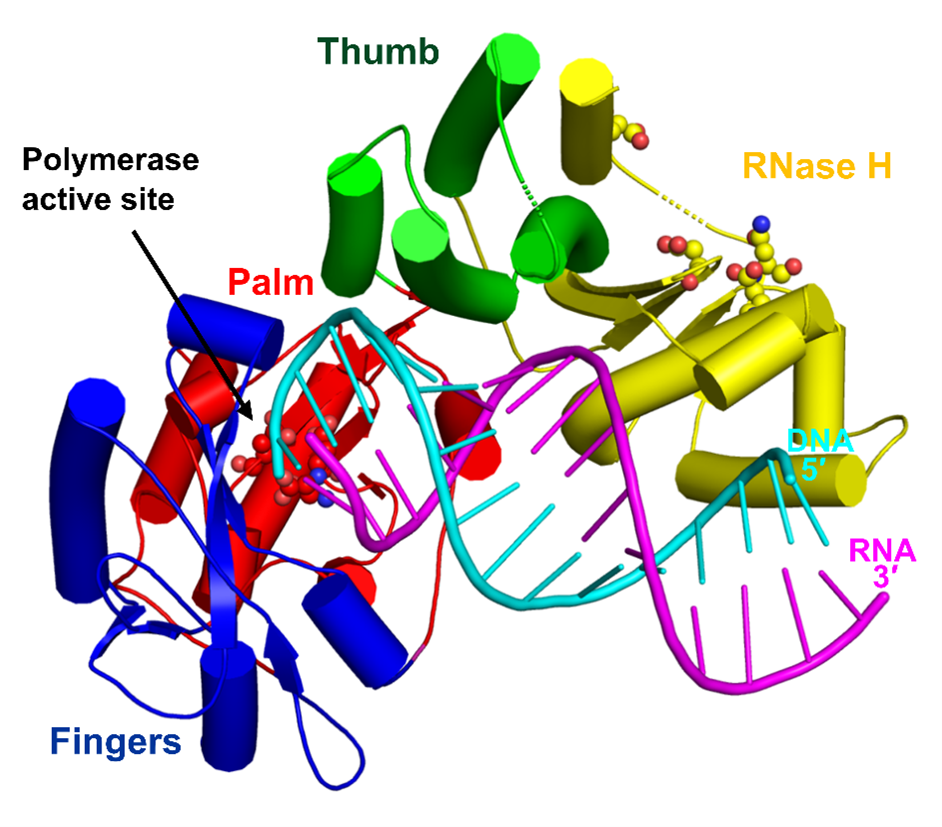
Model of the full length XMRV RT based on SAXS data, a crystal structure of the polymerase-connection fragment in complex with RNA/DNA hybrid (blue-fingers, red-palm, green-thumb, yellow-connection) and the structure of isolated XMRV RNase H domain (orange, Zhou D, J Struct Biol. 2012). RNA template stand is in purple and DNA primer strand in blue.
Foamy viral protease-reverse transcriptase
One of the distinguishing features of foamy viruses (FV) is the different way of replication. Additionally they synthesize a separate mRNA from which the Pol protein is expressed as a precursor of PR-RT-IN, which is later cleaved into two proteins: PR-RT and IN. Thus, FV RT is the only protein of its type to contain an additional PR domain located at the N-terminus of the protein.
Nowacka M*, Nowak E*&, Czarnocki-Cieciura M, Jackiewicz J, Skowronek K, Szczepanowski RH, Wöhrl BM, Nowotny M&. Structures of substrate complexes of foamy viral protease-reverse transcriptase. J Virol., 2021; 95(18): e0084821. *- equally contributing, & - corresponding authors
-
First structures of substrate complexes of a foamy viral protease-reverse transcriptase.
-
Novel mechanism of switching between protein monomer on RNA/DNA substrate and dimer on dsDNA.
-
Mobile RNase H domain of foamy viral PR-RT transiently interacts with the RNA/DNA substrate 18-21 nt from the polymerase active site.
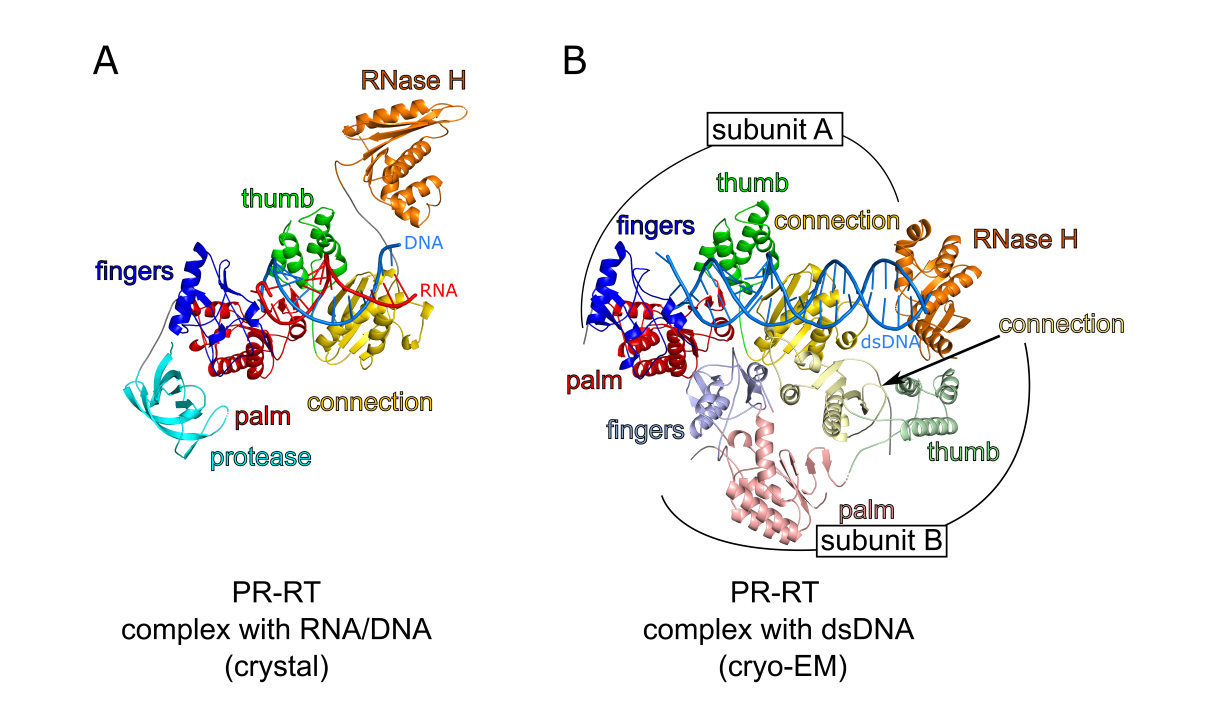
Structures of nucleic acid complexes of MFV PR-RT. (A) Crystal structure of MFV PR-RT in complex with an RNA/DNA hybrid substrate. (B) Cryo-EM structure of MFV PR-RT ΔRH in complex with dsDNA. Lighter shades of colors are used for domains/subdomains of subunit B.
Ty3 reverse transcriptase
Retrotransposons are mobile genetic elements that replicate with an RNA intermediate. Reverse activity of element-encoded RT is essential for this process. Retroelements are one of the most potent forces shaping eukaryotic genomes – more than 40% of human genome derives from those elements. Ty3 is a yeast retrolement from long-terminal class termed Ty3/Copia. It is thought that retroviruses evolved from this class of retrotransposons.
Nowak E, Miller JT, Bona MK, Studnicka J, Szczepanowski RH, Jurkowski J, Le Grice SFJ&, Nowotny M&. Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry. Nature Struct. Mol. Biol., 2014; 21(4):389-96;
& - corresponding authors
-
The first crystal structure of a retrotransposon RT.
-
Ty3 RT forms an asymmetric homodimer in which one subunit has the polymerase competent configuration and the other has an altered conformation and harbors the RNase H activity.
-
RNase H is postulated to undergo a conformational change to reach the position required for RNA/DNA cleavage, which regulates this activity.
-
Ty3 and HIV RT architecture differs: HIV enzyme is a constitutive heterodimer with both polymerase and RNase H activities residing in the larger subunit and Ty3 is a substrate-induced homodimer in which the two activities are located in different subunits.
-
The studies of XMRV and Ty3 reverse transcriptases have been performed in collaboration with Dr. Stuart Le Grice (National Institutes of Health, USA).

Crystal structure of Ty3 reverse transcriptase. The subunit with polymerase-competent configuration is shown in darker color (blue-fingers, red-palm, green-thumb, yellow-RNase H) and the subunit with altered conformation in lighter shades of the same colors. RNA template stand is in purple and DNA primer strand in blue. Active site residues for polymerase and RNase H domain are shown as sticks.
Cauliflower mosaic virus reverse transcriptase (CaMV RT)
CaMV is a prominent plant virus, important in both plant pathology and agricultural biotechnology. Even though the genome of CaMV is a double-stranded DNA, it replicates through reverse transcription. Interestingly, the structural features of CaMV RT make it similar to Ty3 RT, but its mode of action and enzymatic properties differ from other reverse transcriptases.
Chandrasekaran Prabaharan, Małgorzata Figiel, Roman H. Szczepanowski, Krzysztof Skowronek, Weronika Zajko, Vinuchakkaravarthy Thangaraj, Sebastian Chamera, Elzbieta Nowak, and Marcin Nowotny, Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase, J. Biol. Chem. (2024) 300(8), 107555.
-
The first crystal structure of a plant viral RT in complex with an RNA/DNA hybrid substrate.
-
CaMV RT forms a monomeric complex with the hybrid substrate and adopts a polymerase configuration. A single molecule of CaMV RT performs DNA synthesis.
-
Novel mechanism of RNase H activity that requires two copies of the enzyme for RNA hydrolysis. One molecule is involved in structural stabilisation and another forms a transient association with the monomeric complex for RNA cleavage.
Crystal structure ofCaMV RT-RNA/DNA hybrid complex. Domains of CaMV RT are labelled and RNA and DNA strands are shown in magenta and cyan, respectivel
HIV-1 reverse transriptase
HIV-1 RT is an important drug target in therapy of HIV-1 infection. The RT uses its polymerase and RNase H activities to catalyze the process of reverse transcription, in which the single-stranded RNA of the virus is converted into double-stranded DNA that can be integrated into the host cell genome.
Figiel M, Krepl M, Poznański J, Gołąb A, Šponer J, Nowotny M. Coordination between the polymerase and RNase H activity of HIV-1 reverse transcriptase. Nucleic Acids Res., 2017; 45(6):3341-3352.
-
Dynamics of HIV-1 RT-RNA/DNA complex were studied using a combination of chemical cross-linking and molecular dynamics simulations.
-
The RNA/DNA substrate can simultaneously interact with the polymerase and RNase H active sites.
-
Untwisting of the RNA/DNA substrate double helix is required for its productive interaction with the RNase H active site.
-
This allows HIV-1 RT to regulate the amount of the RNase H activity.
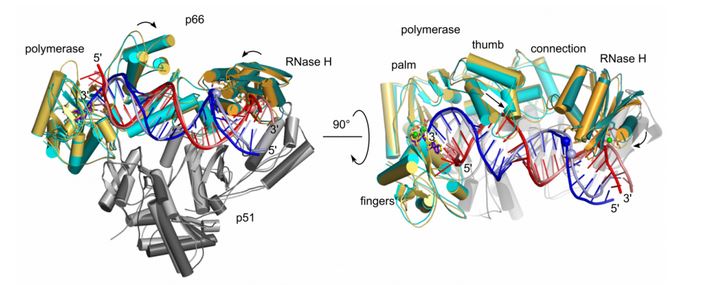
Molecular dynamics simulation of HIV-1 RT RNA/DNA complex (two views). Superposition of the starting model (light colors) and the final model in MD simulations (dark colors). Domains of HIV-1 RT are labeled. RNA and DNA strands of the substrate are shown in red and blue, respectively.
In order to be integrated into the host cell genome, the single-stranded RNA of HIV-1 is converted into double-stranded DNA. The synthesis of the (+)-strand DNA starts from the polypurine tract (PPT) primer. The PPT primer is generated by the RNase H domain of HIV-1 RT which cuts specifically at its termini, but leaves the body of the PPT intact.
Figiel M, Krepl M, Park S, Poznański J, Skowronek K, Gołąb A, Ha T, Šponer J, Nowotny M. Mechanism of polypurine tract primer generation by HIV-1 reverse transcriptase. J Biol. Chem., 2018; 293(1):191-202.
-
Factors involved in recognition of the PPT sequence by HIV-1 RT-RNA/DNA were studied using a combination of chemical cross-linking, molecular dynamics simulations, and single-molecule assays.
-
The PPT is specifically recognized after the complex with HIV-1 RT is formed and not at the stage of binding.
-
Recognition of the PPT is based on two elements: agreement with the sequence preference of RNase H and the indirect readout of the poly-rA/dT stretch. The rigid but brittle poly-rA/dT is not compatible with the catalytically relevant substrate geometry and is prone to undergo sequence slippage upon deformation.
-
The poor match with the RNase H sequence preference and the dynamic properties of the PPT explain the protection of its body from cleavage.
Studies of HIV-1 RT have been done in collaboration with Jiří Šponer (Institute of Biophysics, CAS) and Taekjip Ha (Johns Hopkins Univ.).
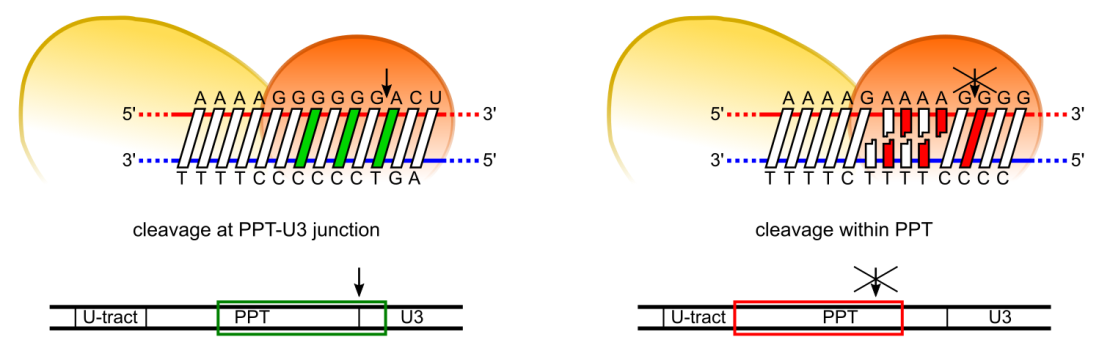
Model of PPT recognition by HIV-1 RT. Cleavage at the expected site involves both preferred residues at the cleavage consensus position and the ability of PPT sequence to undergo the appropriate conformational change without distortion (left panel). Cleavage in the middle of the PPT body (A-tract) is inhibited by three elements: non-preferred residues in the consensus positions, rigidity of the poly(rA-dT) sequence, and misalignment of the substrate at the RNAse H active site due to poly(rA-dT) sequence slippage
Abi polymerases
Abortive infection (Abi) is a bacterial antiphage defense strategy involving suicide of the infected cell. Some Abi pathways involve polymerases that are related to reverse transcriptases and combine the ability to synthesize DNA in a template-independent manner with protein priming. The first reverse transcriptase-related proteins that were proposed to utilize the abortive infection antiphage strategy were AbiK, Abi-P2 and AbiA. Within this set, AbiA is the only protein to contain an additional component – a higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain which is also found in several other systems of antiviral defense.
Figiel M*&, Gapińska M&, Czarnocki-Cieciura M, Zajko W, Sroka M, Skowronek K, Nowotny M*. Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases. Nucleic Acids Res., 2022; 50(17):10026-10040.
& - equally contributing; * - corresponding authors
-
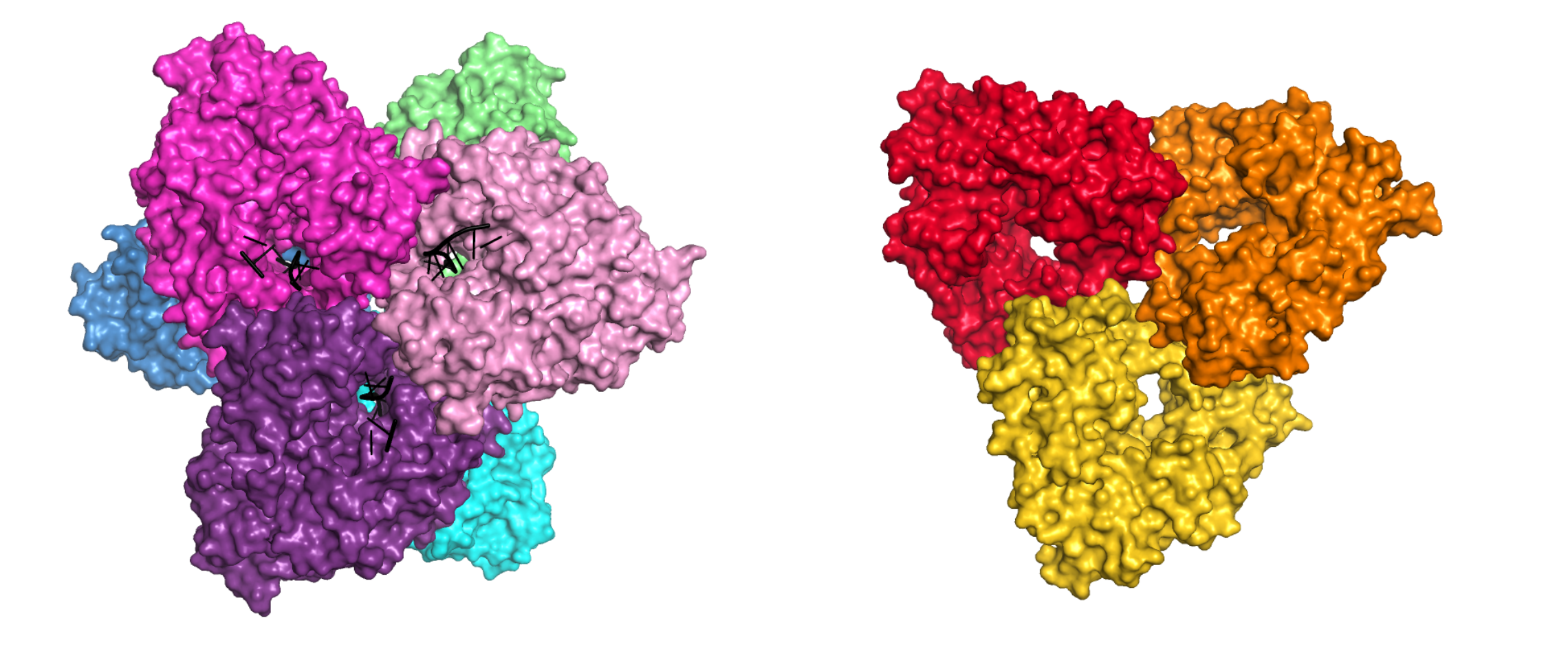
Cryo-EM and crystal structure of AbiK-DNA adduct, cryo-EM structure of protein-priming deficient AbiK variant, crystal structure of Abi-P2
-
AbiK-DNA structure is the first one showing a protein-DNA adduct resulting from protein-primed DNA synthesis
-
AbiK and Abi-P2 are the first examples of RT-related enzymes that adopt hexameric or trimeric architecture; the oligomerization is required for enzymatic activity
-
AbiK and Abi-P2 adopt a bilobal structure with an RT-like domain that comprises palm and fingers subdomains and a unique helical domain
-
Protein priming by Ll-AbiK involves a conformational change of a mobile loop harboring the priming tyrosine residue
Crystal structures of AbiK-DNA adduct (left) and Abi-P2 (right) polymerases shown in surface representation. Individual subunits are shown in different colors, DNA strands are shown as black cartoon.
Gapińska M, Zajko W, Skowronek K, Figiel M, Krawczyk PS, Egorov AA, Dziembowski A, Johansson MJO*, Nowotny M*. Structure-functional characterization of Lactococcus AbiA phage defense system. Nucleic Acids Res., 2024, 52(8):4723-4738.
* - corresponding authors
-
a crystal structure of AbiA-DNA complex
-
AbiA forms dimers and HEPN domain is essential for dimerization
-
AbiA exhibits template-independent DNA polymerase activity with nucleotide preference for adenosines and cytosines
-
HEPN domain is enzymatically inactive, yet its presence is required for DNA polymerase activity of AbiAProtein priming by Ll-AbiK involves a conformational change of a mobile loop harboring the priming tyrosine residue
-
DNA polymerase activity of AbiA is essential for protection against invading phages as evidenced by an infection assay in E. coli surrogate host
-
contrary to previous assumptions, the antiphage defense mediated by AbiA and AbiK appears not to rely on induced cell death
Studies of AbiA were performed in collaboration with Marcus Johansson (Lund University, Sweden) and Andrzej Dziembowski (IIMCB).
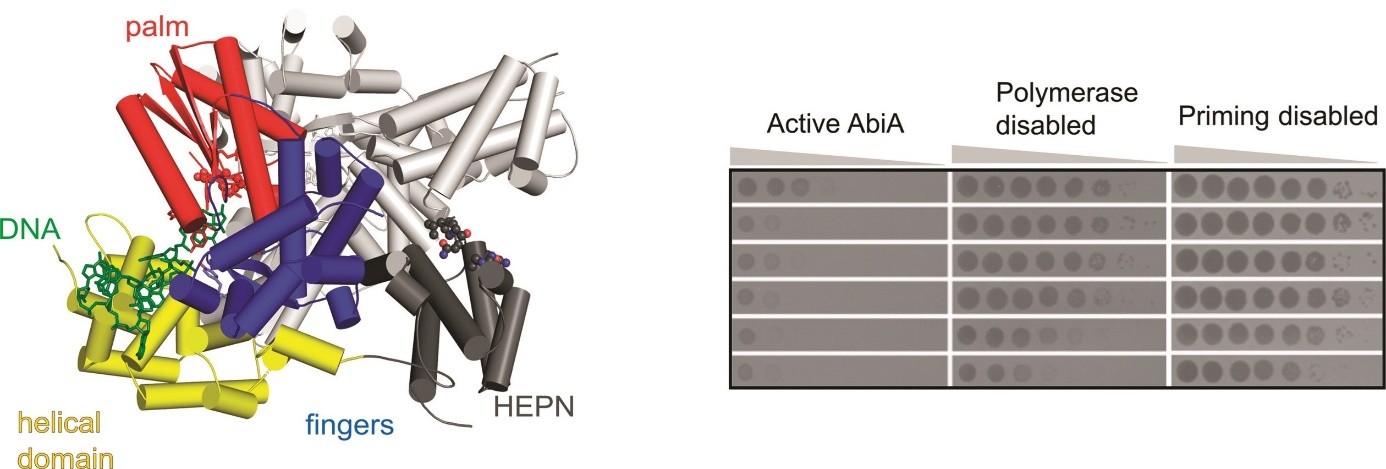 (left) Crystal structure of L. lactis AbiA protein dimer. Domains of one of the subunits are colored and labeled, DNA product of the enzyme is shown in green.
(left) Crystal structure of L. lactis AbiA protein dimer. Domains of one of the subunits are colored and labeled, DNA product of the enzyme is shown in green.
(right) Results of the infection assay, in which E. coli cells harboring wild-type and mutated versions of AbiA were challenged by different phages. Formation of plaques indicates successful infection.