DNA repair
Bacterial Nucleotide Excision Repair
Nucleotide excision repair (NER) is one of the major pathways of DNA repair and its main feature is its ability to recognize a wide spectrum of DNA lesions of various sizes and structures. In bacterial NER UvrA, a dimeric ATPase, plays the role of a DNA damage sensor. Damage verification is performed by UvrB helicase and UvrC double nuclease excises the damaged DNA fragment.
Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nature Struct. Mol. Biol., 2011; 18:191-197
-
The first crystal structure of UvrA interacting with damaged dsDNA obtained using the protein from T. maritima.
-
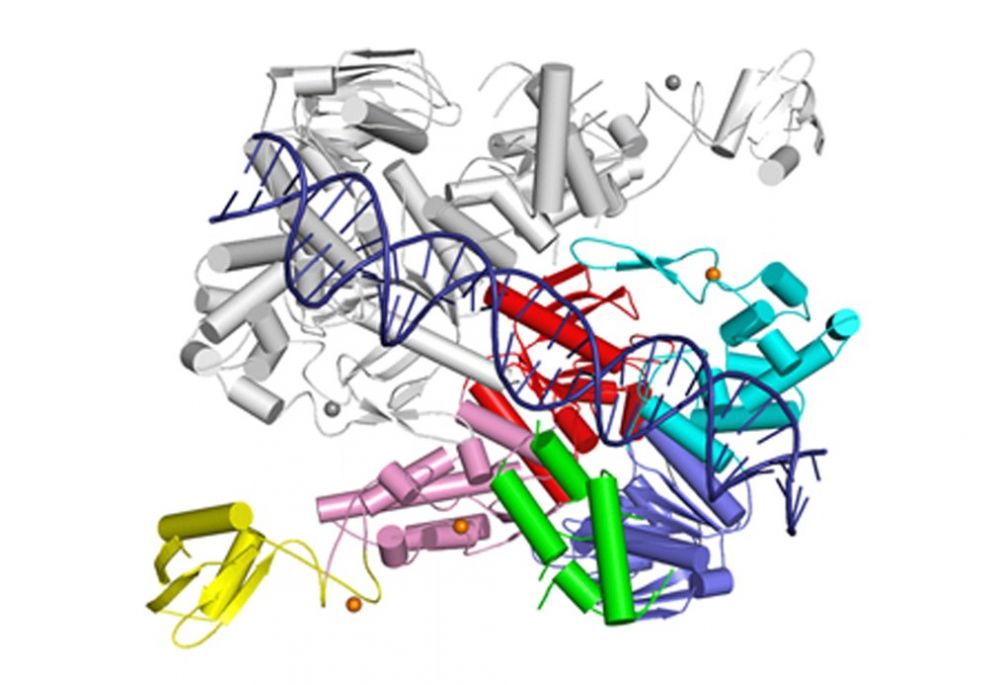
UvrA does not interact with the damage site directly but senses the deformed conformation of the DNA induced by the presence of the lesion – bending, stretching and unwinding.
Crystal structure of UvrA dimer (one monomer in color, the other in gray). One ATPase module is shown in cyan and red and the other in pink and blue. The DNA-binding domain is in green and UvrB-binding domain in yellow. Structural zinc ions are shown as orange spheres.
Jaciuk M*, Swuec P*, Gaur V*, Kasprzak JM, Renault L, Dobrychłop M, Nirwal S, Bujnicki JM& , Costa A&, Nowotny M&. A combined structural and biochemical approach reveals translocation and stalling of UvrB on the DNA lesion as a mechanism of damage verification in bacterial nucleotide excision repair. DNA Repair, 2019; 85, 102746; * - equally contributing, & - corresponding authors
-
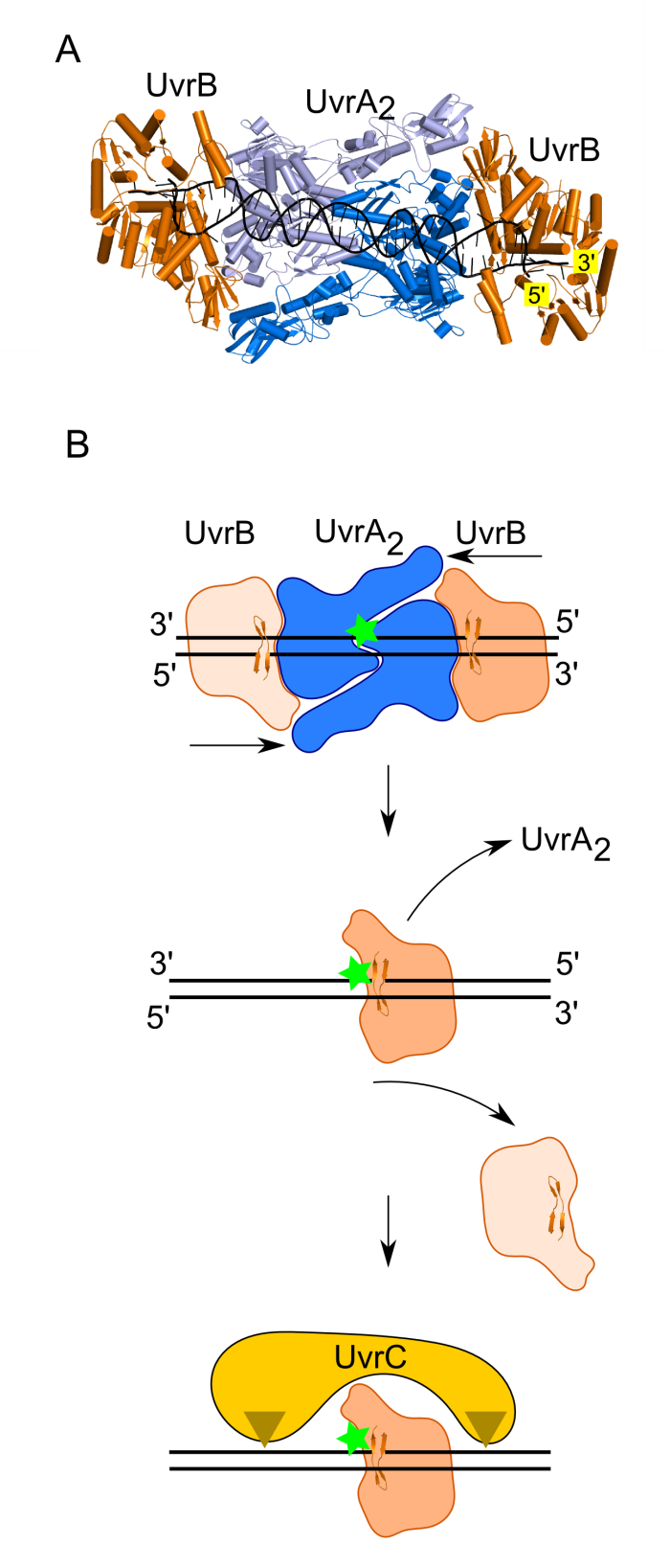
Computational structural model of UvrA—UvrB—DNA complex involved in DNA damage verification in bacterial DNA repair was prepared and corroborated experimentally by electron microscopy.
-
UvrB uses a β-hairpin element to clamp one DNA strand, each one of the two UvrB molecules in the complex clamps a different DNA strand.
-
UvrB translocates in 3′ direction toward the DNA lesion where the UvrB molecule clamping the damaged strand stalls and recruits UvrC nuclease.
-
This mechanism explains how the initial imprecise localization of the damage by UvrA is converted to precise and strand-specific localization to promote accurate incisions by UvrC.
The studies of damage verification in bacterial NER have been performed in collaboration with Janusz M. Bujnicki (IIMCB) and Alessandro Costa (The Crick Institute).
Model of damage verification in bacterial NER. (A) Model of UvrA2—UvrB2—DNA complex. UvrA dimer shown in two shades of blue. UvrB shown in orange. DNA shown in black. (B) Proposed mechanism. The UvrA dimer bound at the site of DNA modification recruits two UvrB molecules. Each UvrB molecule clamps a different DNA strand under the B-hairpin element (upper panel). Both UvrB molecules then translocate toward the lesion with 5′ to 3′ polarity on the strand under the hairpin. The UvrB molecule that clamps the modified strand will stall at the lesion (green star indicates the site of DNA modification) and the other UvrB molecule (light orange) will dissociate (middle panel). The stalled UvrB recruits UvrC double nuclease (shown in yellow), which makes two incisions indicated with triangles.
Rad2
Eukaryotic nucleotide excision repair (NER) is one of the major DNA repair pathways. It involves the excision of the DNA fragment containing the damage. This is achieved through the action of two nucleases – XPF-ERCC1 complex and XPG (Rad2 in yeast). Rad2/XPG belongs to flap endonuclease metal ion-dependent enzymes along with FLAP1 and EXO1. Its unique feature within this family is the ability to cleave DNA bubbles – substrates with melted single stranded region flanked with double-stranded stretches of DNA.
Miętus M, Nowak E, Jaciuk M, Kustosz P, Studnicka J, Nowotny M. Crystal structure of the catalytic core of Rad2: insights into the mechanism of substrate binding. Nucleic Acids Res., 2014; 42(16):10762-75.
-
The first crystal structure of the catalytic core of Rad2 using a truncated version of the S. cerevisiae enzyme.
-
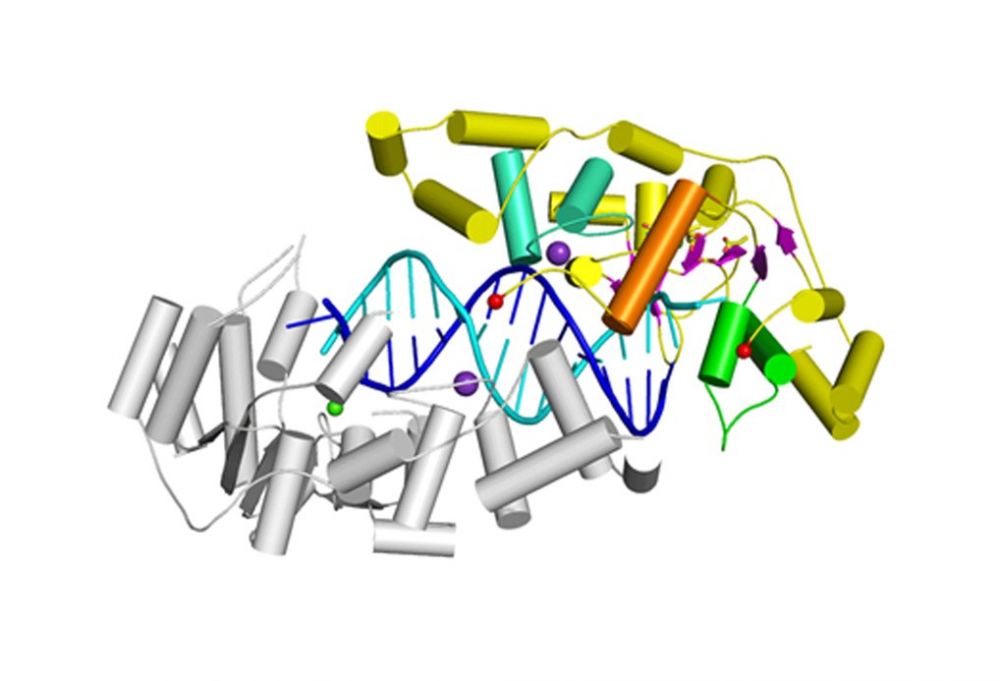
The main substrate specificity determinant of Rad2 is the interaction of the last exposed base pair of the double-stranded region with the so-called hydrophobic wedge of the enzyme. No interactions with the single stranded portion of the substrate are observed.
-
The main DNA-binding element is potassium-coordinating helix-two-turn-helix (H2TH) motif. It contains an additional charged helix binding the DNA, which is a unique feature of Rad2.
-
The likely explanation for the unique ability of Rad2 to clave DNA bubbles (substrate without a free 5’ DNA end) is the altered structure of the so-called helical arch. In FEN1 and EXO1 this element blocks the exit from the active site preventing cleavage of substrates without a free 5’ end. In Rad2 the helical arch has a different structure forming an exit route from the active site.
Crystal structure of Rad2-DNA complex. The complex contains two independent protein molecules – one is shown in color: cyan for H2TH motif, green for hydrophobic wedge and orange for helical arch. The DNA is shown in cyan and blue, potassium ion as a purple sphere and the calcium ion at the active site as a green sphere.
RecFOR
RecFOR pathway is one of the homologous recombination-based DNA repair pathways in bacteria. It involves RecF, RecO and RecR proteins, that bind at the junction of single-stranded (ss) and double-stranded (ds) DNA and then facilitate the replacement of the SSB protein, which initially covers ssDNA, with RecA which promotes the search for the homologous sequence.
Nirwal S, Czarnocki-Cieciura M, Chaudhary A, Zajko W, Skowronek K, Chamera S, Figiel M, Nowotny M. Mechanism of RecF–RecO–RecR cooperation in bacterial homologous recombination. Nature Structural & Molecular Biology, 2023; doi: 10.1038/s41594-023-00967-z.
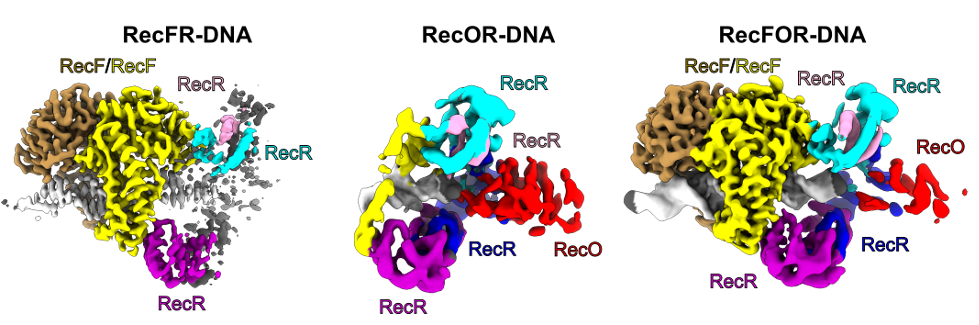
- The structure of RecF-DNA complex at 3.1 Å resolution, shows that RecF dimer uses its helical protrusions to clamp the dsDNA.
- The structure of RecF-RecR-DNA subcomplex (3.1 Å resolution), shows that one protomer of RecF dimer binds two different regions of the tetrameric RecR ring.
- The structures of RecR-RecO-DNA complex (6.1 Å resolution) and RecF-RecO-RecR-DNA assembly, explains how RecO is positioned to interact with ssDNA and SSB, which is proposed to lock the complex at a ss-dsDNA junction.
The cryo-EM reconstructions for the RecFOR-DNA assembly and the RecFR-DNA and RecOR-DNA subcomplexes of the assembly. RecF dimer is shown in yellow and sand; RecR tetramer in cyan, pink, purple and blue; RecO in red and DNA in white.
RuvC
RuvC is a canonical bacterial Holliday junction (HJ) resolvase, which functions as a dimer. HJ are four-way DNA structures formed by the exchange of strands between two helices. They are intermediates in homologous recombination, a process which is used to repair dangerous DNA lesions such as double-strand breaks.
Gorecka, KM, Komorowska W and Nowotny M. Crystal structure of RuvC resolvase in complex with Holliday junction substrate. Nucleic Acids Res., 2013; 41(21):9945-55
-
The first crystal structure of RuvC in complex with a DNA substrate and the first substrate complex structure of a cellular resolvase, solved at 3.8 Å resolution.
-
HJ in a novel tetrahedral conformation with two phosphate groups symmetrically located 1 nt from the HJ exchange point interacting with two active sites of RuvC dimer.
-
Novel mode of HJ recognition relative to phage enzymes for which complex crystal structures had been available.
Crystal structure of RuvC in complex with Holliday junction. The two protomers are shown in pink and orange. The DNA is in blue with cleaved phosphates indicated with spheres.
Górecka KM*, Krepl M*&, Szlachcic A, Poznański J, Šponer J, Nowotny M&. RuvC uses dynamic probing of the Holliday junction substrate to achieve sequence specificity and efficient resolution. Nature Commun. 2019; 10(1):4102; *- equally contributing,
& - corresponding authors
-
Mechanism of sequence specificity and cut coordination revealed by a combination of structural biology, biochemistry, and a computational approach.
-
Correct positioning of the substrate for cleavage requires conformational changes within the bound DNA, which are possible only for the cognate sequence.
-
The conformational changes and the relieving of protein-induced structural tension of the DNA facilitates coordination between the two cuts.
-
The unique DNA cleavage mechanism of RuvC demonstrates the importance of high-energy conformational states in nucleic acid readout.
Studies of RuvC mechanism have been done in collaboration with Jiří Šponer (Institute of Biophysics, CAS)
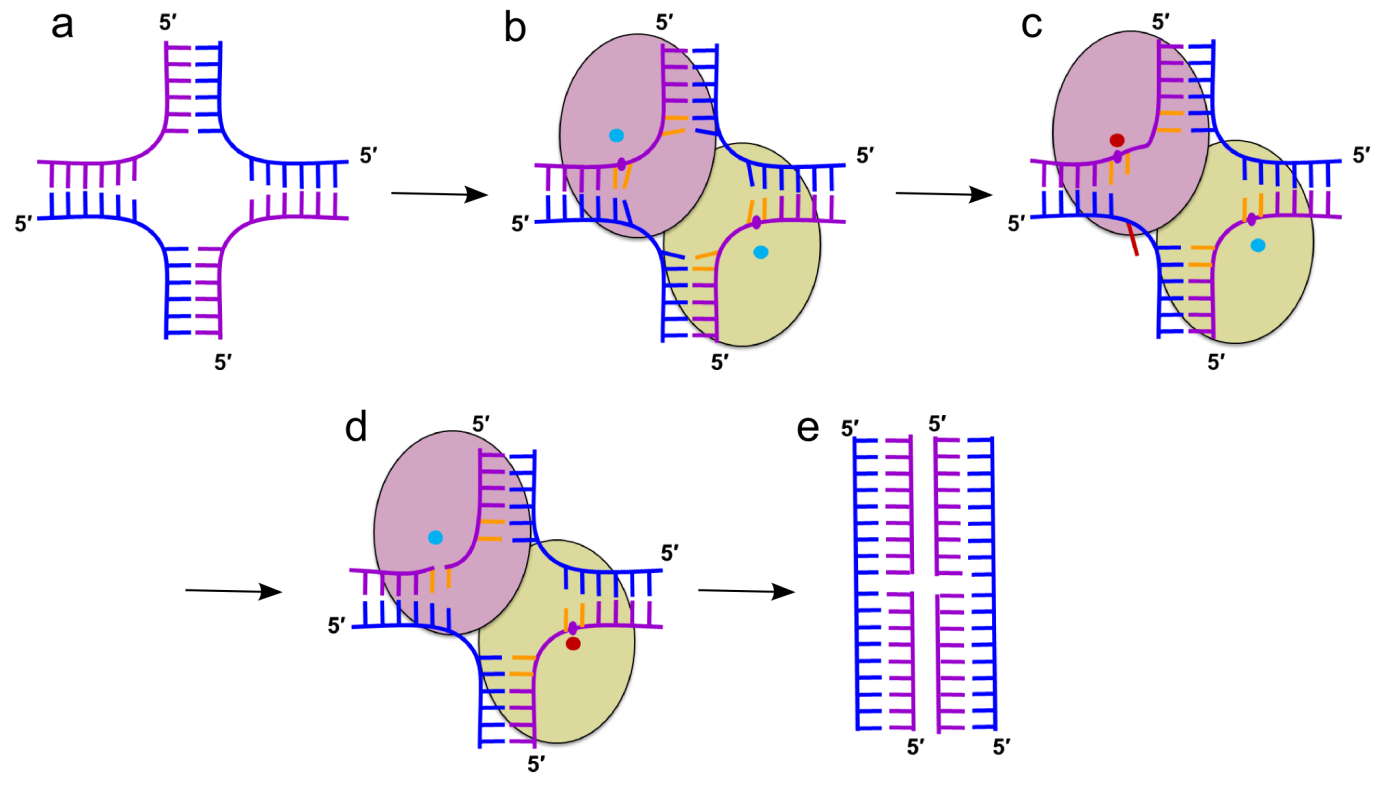
Cartoon representation of the mechanism of HJ resolution by RuvC. (a) Holliday junction. Cleaved and non-cleaved DNA strands are shown in purple and blue ladder-like representations, respectively. (b) Binding of the HJ DNA. The subunits of the dimer are shown as yellow-green and pink ovals. The scissile phosphate is marked as a purple circle. Cyan circles show active sites in an inactive configuration. (c) Flipping of the adenine (red) opposite the scissile base. The active site in the catalytic configuration is shown as a red circle. (d) The second cut. (e) Resolution products
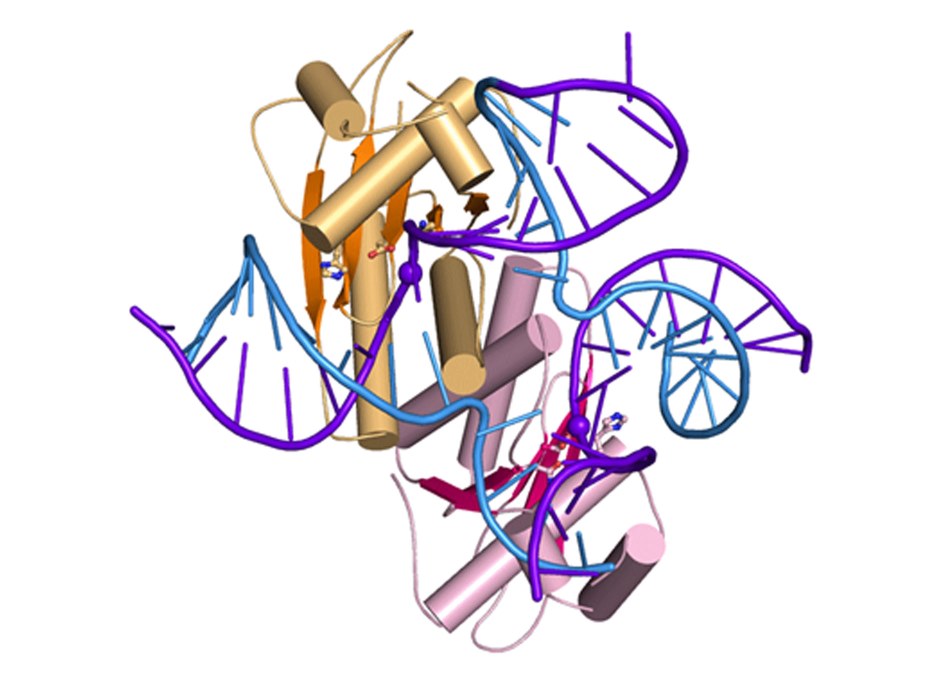
Slx1-Slx4
Slx1 is a nuclease which cleaves various DNA structures during DNA repair and recombination. It associates with Slx4 platform protein which coordinates the action of multiple proteins. Slx1 together with Mus8-Eme1 nuclease constitute one of the major Holliday junction pathways in eukaryotes.
Gaur V, Wyatt HD, Komorowska W, Szczepanowski RH, de Sanctis D, Gorecka KM, West SC, Nowotny M. Structural and Mechanistic Analysis of the Slx1-Slx4 Endonuclease. Cell Rep., 2015; S2211-1247(15)00165-5
-
First structural information for Slx1 and Slx4CCD.
-
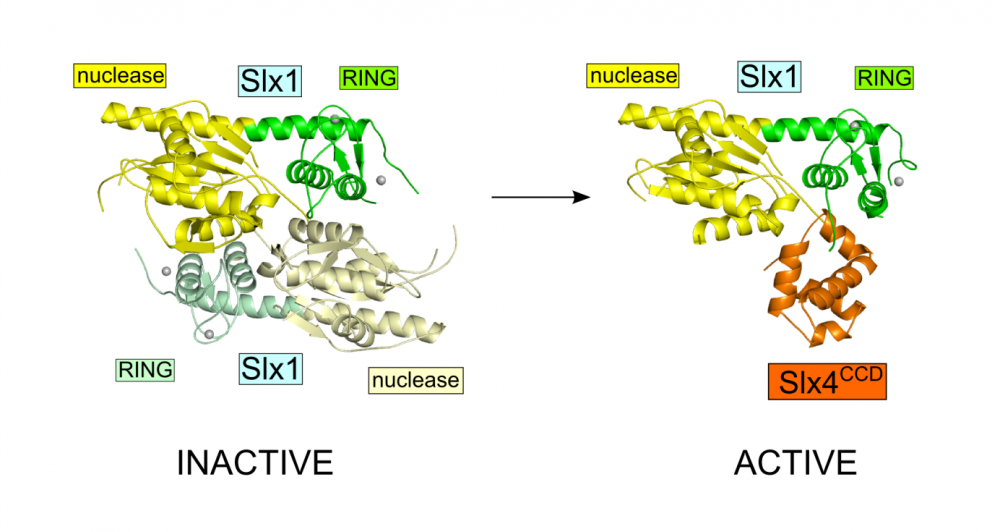
Fungal Slx1 forms a homodimer in which the active site is blocked, explaining why Slx1 alone is inactive.
-
Slx4CCD domain binding is mutually exclusive with homodimerization.
-
Slx4 binding exposes the active site of Slx1 and activates the nuclease. This mechanism ensures that the promiscuous and potentially dangerous Slx1 nuclease is only active when bound to Slx4 platform which regulates its activity and coordinates it with other proteins.
Crystal structures of Slx1 homodimer and Slx1 in complex with Slx4CCD domain (orange). Slx1 comprises GIY-IYG nuclease domain (yellow) and RING finger zinc-binding domain (green). Upon Slx4 binding the active site of the nuclease domain is exposed and the enzyme is activated.
Gaur V, Zajko W, Nirwal S, Szlachcic A, Gapińska M, Nowotny M. Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease. Nucleic Acids Res., 2019; 47:11681-11690
-
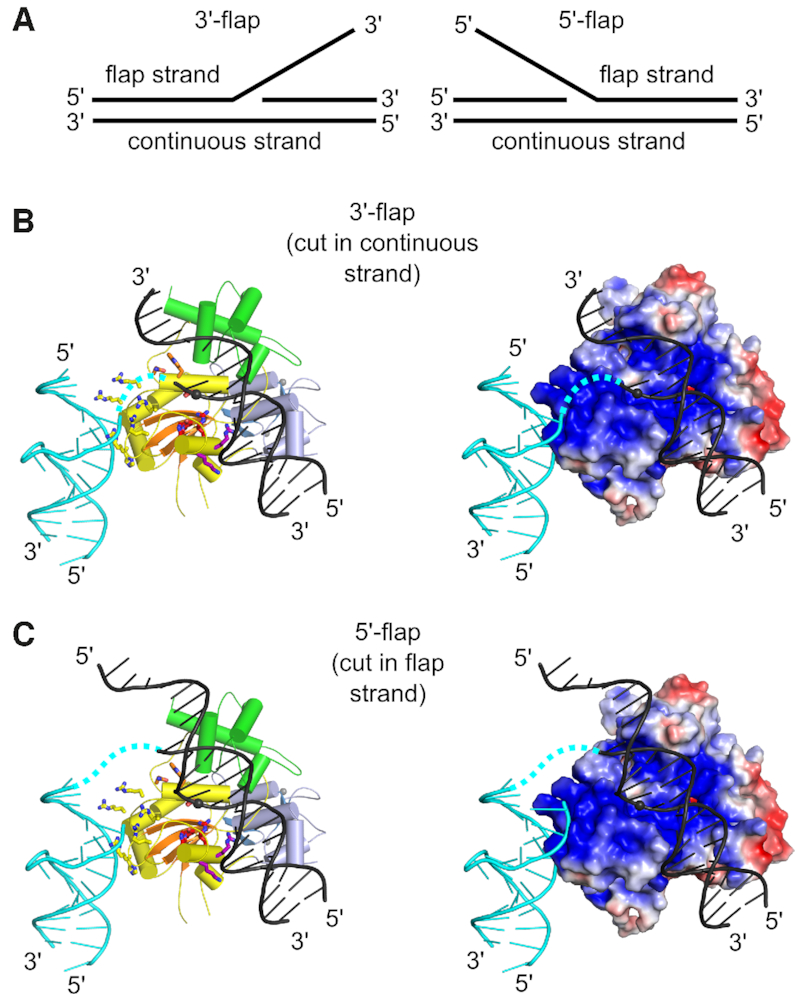
Based on a crystal structure, modeling, and biochemical studies, a mechanism was proposed to explain the specificity of Slx1 towards a wide range of branched DNA substrates.
-
Slx1 bends the DNA and identifies the branch point as a flexible discontinuity.
-
Cuts are introduced on the 3ʹ side of the branch point.
Models of Slx1–Slx4CCD3 interactions with branched DNA. (A) Schematic of the DNA flap substrates with terminology of the strands. (B) Model of Slx1–Slx4CCD3 bound to a 3′-flap substrate in configuration, which is conducive to incision in the continuous strand. (Left) Slx1 GIY-YIG domain is shown in yellow with -strands in orange and RING domain shown in blue. Slx4CCD is shown in green. The modeled DNA is based on the structure of R.Eco29kI restrictase (PDB ID: 3NIC) and is shown in black with the scissile phosphate shown as a sphere. A fragment of the DNA that is observed in the Slx1–Slx4CCD-DNA structure is shown in cyan. The possible link between the two DNA double helices is shown as a dashed cyan line. Residues of the active site are shown as red sticks. The same model with protein in surface representation, colored according to the surface potential (±3 Kt/e). (C) Model of Slx1–Slx4CCD3 bound to a 5′-flap substrate, in configuration which is conducive to incision of the flap strand. The representations are the same as in (B).