DNA transposition
Tn7 Transposase TnsB
Bacterial Tn7 elements are among the best-studied and most widespread DNA transposons and are detected in 10%–20% of sequenced bacterial genomes. Tn7 mobility is tightly controlled and mediated by five element-encoded proteins. Translocation of the Tn7 is executed by a heteromeric transposase TnsA+TnsB. The TnsB transposase recognizes the left and right ends of the element, catalyzes cleavage at the 3′ ends of the transposon, and performs the strand-transfer reaction that leads to the insertion of the element into a target DNA.
Kaczmarska Z*, Czarnocki-Cieciura M*, Górecka-Minakowska KM*, Wingo RJ, Jackiewicz J, Zajko W, Poznański JT, Rawski M, Grant T, Peters JE&, Nowotny M.& Structural basis of transposon end recognition explains central features of Tn7 transposition systems. Mol Cell, 2022 Jul 21;82(14):2618-2632.e7
* - equally contributing, & - corresponding authors
-
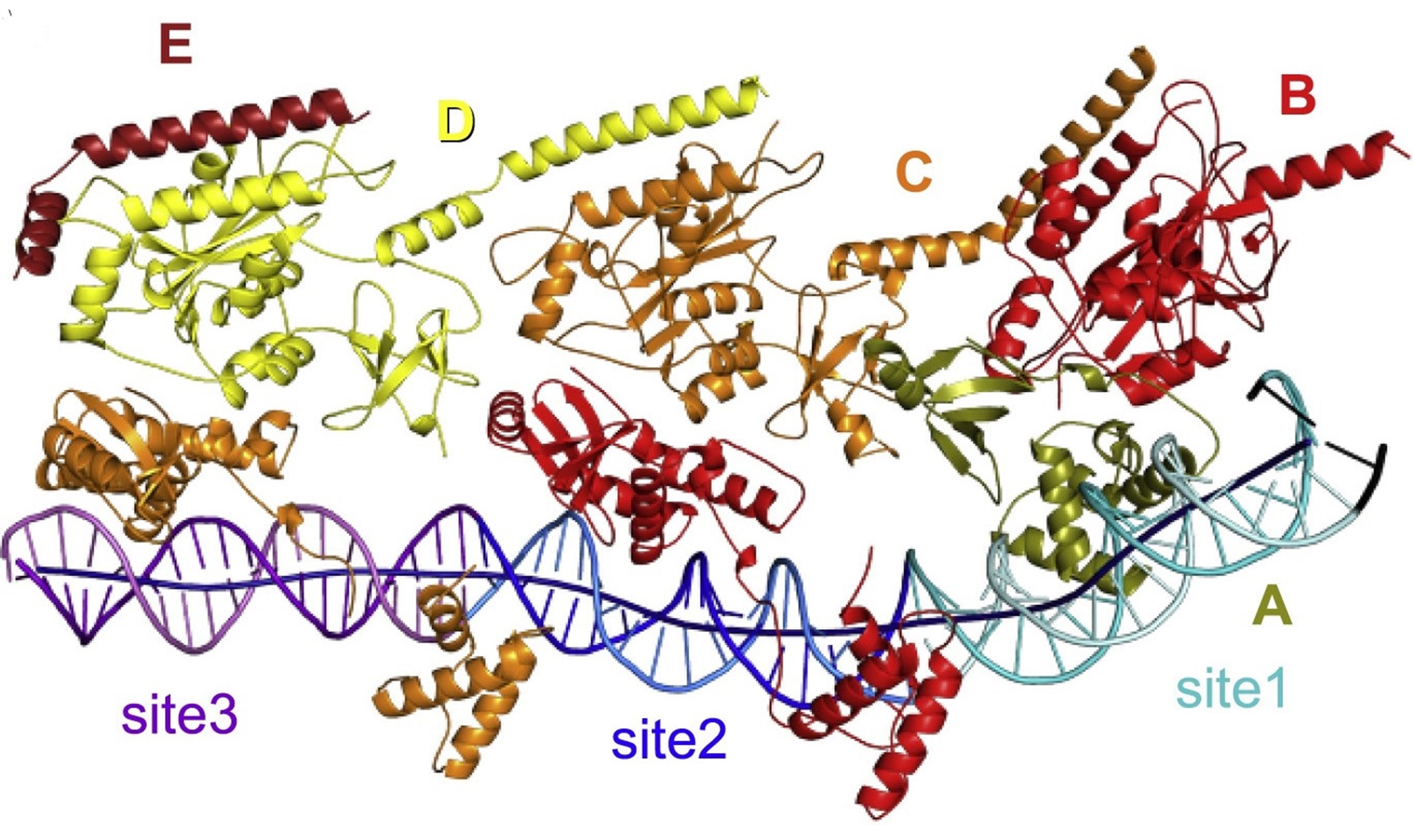
Cryo-EM structure of Tn7 transposase TnsB in complex with transposon end DNA
-
TnsB chains interact with its binding sites in DNA in a tiled and intertwined fashion
-
TnsB exhibits a DNA sequence preference rather than strict specificity
-
Overlap of TnsB-binding sites in the right Tn7 end is key for its functionality
Beads-on-a-string organization of TnsB bound to the fragment of the right transposon end. The TnsB chains (labeled with capital letters) and DNA sites are colored as follows: olive for chain A, red for chain B, orange for chain C, yellow for chain D, burgundy for chain E, light cyan/cyan (top/bottom strands) for site 1, blue/dark blue for site 2, pink/purple for site 3, and black for transposon terminus. The axis of the DNA helix is shown as a dark blue tube.