RNases H
Bacterial RNase H2
RNase H2 cleaves the 5′ phosphate of ribonucleotides in RNA-DNA junctions. It is the only known enzyme that can initiate the process of mutation-free removal of single ribonucleotides from genomic DNA. Such ribonucleotides are very often misincorporated by replicative polymerases and lead to genomic instability.
Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal Structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol. Cell, 2010; 40:658-670
-
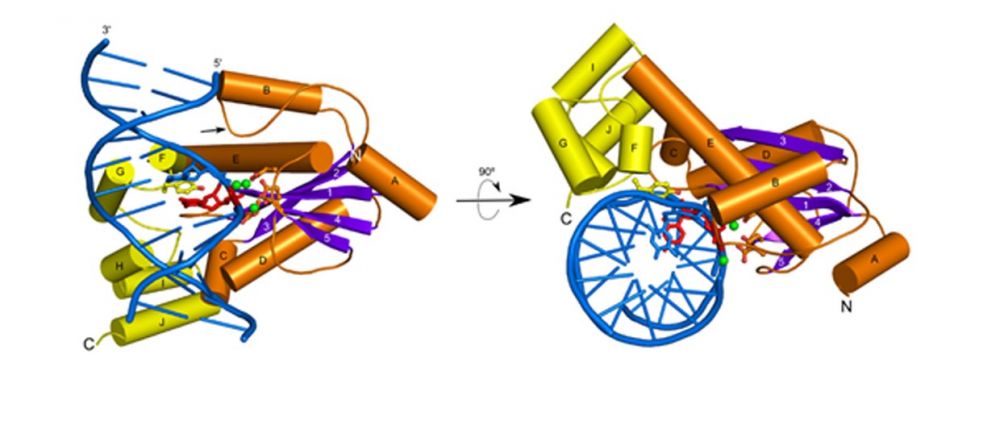
First crystal structure of substrate complex of RNase H2 obtained using T. maritima protein.
-
Unique catalytic mechanism in which the substrate is deformed, so that the phosphate group of the RNA-DNA junction participates in metal ion coordination at the active site. This metal ion positions and activates the nucleophilic water molecule. Hence, the substrate is used to assemble the active site to promote its own cleavage.
The studies of RNases H2 were performed in collaboration with Dr. Robert Crouch (National Institutes of Health, USA).
Crystal structure of T. maritima RNase H2 in complex with the nucleic acid substrate. The catalytic domain is in orange and purple and the helical C-terminal extension in yellow. DNA is shown in blue and single ribonucleotide in red. Metal ions at the active site are shown as green spheres.
Human RNase H2
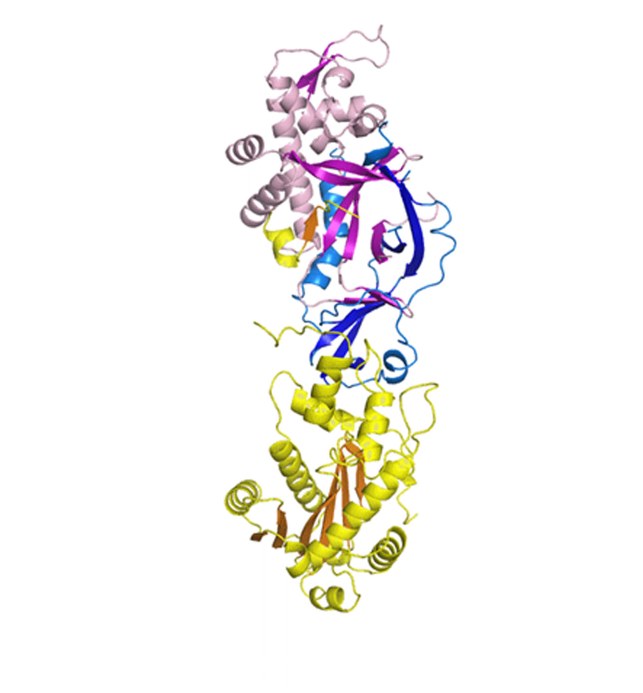
Human RNase H2 comprises three subunits – the catalytic subunit is very similar to monomeric bacterial and archaeal enzymes while the remaining two subunits do not share sequence similarity with any other known proteins. Mutations in human RNase H2 lead to a severe genetic autoimmune disease - Aicardi-Goutières syndrome.
Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects. J. Biol. Chem., 2011; 286:10540-50
-
Crystal structure of the trimeric human RNase H2.
-
All mutations from AGS patients mapped and their effect on the structure and mechanism of the enzyme proposed.
Crystal structure of human RNase H2. The catalytic subunit is shown in yellow and the auxiliary ones in purple in blue.
RNase H3
RNases H3 are present in certain bacterial and archaeal species. They share high sequence and structure similarity with RNases H2, however in terms of biochemical properties they are similar to RNases H1 – they prefer to cleave RNA/DNA hybrids in the middle of the RNA sequence. RNase H3 contains a unique N-terminal domain related to TATA-binding protein.
Figiel M, Nowotny M. Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition. Nucleic Acids Res., 2014; 42(14):9285-94
-
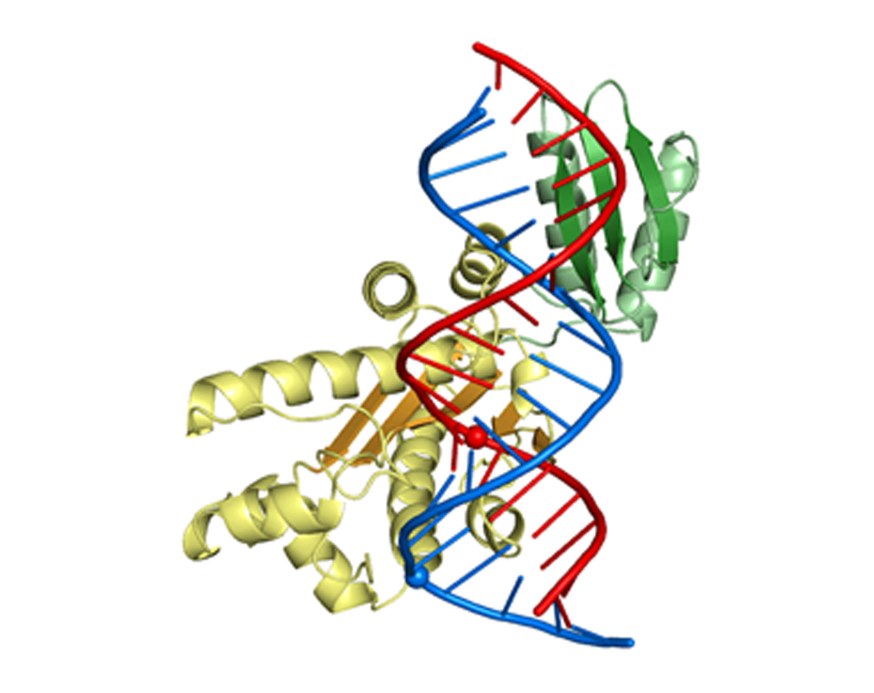
The first crystal structure of RNase H3 in complex with the RNA/DNA substrate.
-
The RNA strand is recognized by contacts between 2’-OH groups of four consecutive ribonucleotides.
-
The DNA strand is recognized by deformation to B-form sugar puckers only allowed for DNA.
-
The N-domain specifically binds RNA/DNA hybrid by recognizing 2’-OH of RNA and forming stacking interactions with the ribose rings of the DNA.
-
The mechanism of N-domain is very similar to the structurally unrelated hybrid-binding domain present in the N-terminus of RNases H1. This is a likely case of convergent evolution of RNA/DNA recognition.
Crystal structure of RNase H3 (catalytic domain in yellow and N-domain in green) interacting with an RNA/DNA hybrid (RNA in red and DNA in blue). The cleaved phosphate is shown as a red sphere and the phosphate group of the deformed DNA residue as a blue sphere.