Our lab investigates the molecular choreography of post-transcriptional gene regulation in bacteria, focusing on the critical moments when these networks are challenged by stress or viral attack. Our research spans from the molecular level, where we use single-molecule microscopy to visualize the assembly of protein-RNA complexes in real time, to the cellular level, where we use bacteriophages to identify novel antibacterial strategies. This integrated approach is applied to commensal and pathogenic E. coli as well as high-priority pathogens like antibiotic-resistant Acinetobacter baumannii.
Research Summary
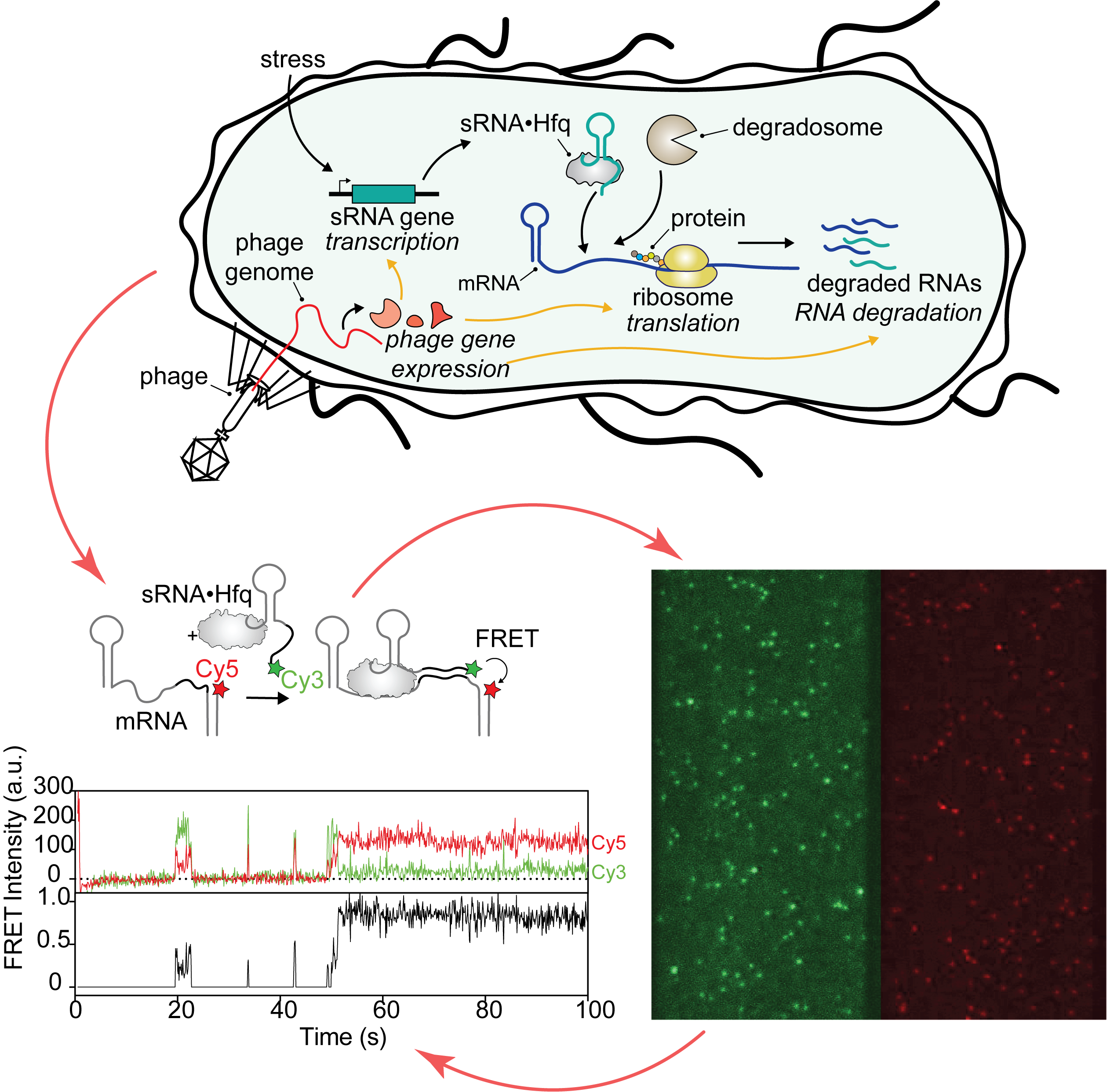
Our research program deciphers the rules of bacterial gene expression across multiple scales. At the molecular level, we use biochemical methods and advanced single-molecule TIRF microscopy to dissect how small regulatory RNAs and the RNA chaperone Hfq function. By capturing real-time interactions between proteins, ribosomes, and RNAs, we seek to understand the fundamental principles governing bacterial gene expression. We then bridge this deep mechanistic insight to the systems level by studying the effects of bacteriophage infection. Here, we deploy transcriptomic and proteomic studies to reveal novel vulnerabilities that can be potentially targeted for antibacterial design. Our work is supported by a Sonata Bis grant from the National Science Centre, Poland and EMBO Installation Grant.
Scientific Impact
- Mechanistic insights: our work provides a mechanistic understanding of how sRNAs select targets and orchestrate gene silencing. By dissecting the dynamics of targeting and degradation, we move beyond static models to reveal fundamental principles of regulatory control in bacteria.
- Cutting-edge technology: we are using a state-of-the-art microscope to monitor single molecules in action. RNA targeting, translation, and degradation can be visualized simultaneously in real time.
- Potential applications: our work on phage-encoded factors targeting antibiotic-resistant bacteria may unlock new antimicrobial strategies against ESKAPE pathogens. Understanding sRNA design rules will enable the development of programmable bacterial regulators for synthetic biology, metabolic engineering, and targeted therapeutic interventions.
Future Goals
Looking ahead, our research will bridge fundamental discovery with therapeutic innovation. We aim to expand our single-molecule analysis to capture the entire regulatory journey of an mRNA, from its initial targeting by sRNA-Hfq complexes to its ultimate fate at the ribosome. We will leverage this deep mechanistic insight to dissect the phage-host arms race in more clinically relevant contexts, such as within ESKAPE pathogens.
Collaborations
We collaborate with Prof. Sander Granneman (University of Edinburgh) to integrate single-molecule visualization with in vivo protein-RNA mapping, and with Prof. Ben Luisi (University of Cambridge) to connect regulatory dynamics to the mechanics of RNA degradation.
Comment
"The central challenge of modern biology is to understand how molecular processes are interconnected at the molecular level within the cellular environment. We're tackling this by visualizing individual RNAs and proteins as they orchestrate bacterial gene regulation – a system of incredible precision and speed. Watching these decisions unfold one molecule at a time reveals the elegant strategies that have ensured bacterial survival for billions of years," says Ewelina Małecka.
Visit the laboratory website for more details: https://maleckalab.com/
 |
Ewelina Małecka, PhD
Correspondence address:
Laboratory of Prokaryotic Gene Regulation
International Institute of Molecular and Cell Biology in Warsaw
4 Ks. Trojdena Street, 02-109 Warsaw, Poland
https://maleckalab.com/
Email: This email address is being protected from spambots. You need JavaScript enabled to view it.
|
DEGREES
2017 - PhD in Biochemistry with Prof. Mikołaj Olejniczak, Adam Mickiewicz University, Poznań, Poland
2012 - MSc in Biotechnology with Prof. Zofia Szweykowska-Kulińska, Adam Mickiewicz University, Poznań, Poland
PROFESSIONAL EXPERIENCE
2022 - present - Head of Laboratory of Prokaryotic Gene Regulation (former: Single-Molecule Biophysics), International Institute of Molecular and Cell Biology in Warsaw, Poland
2022 - present - Visiting Researcher, Dept. of Biochemistry, Johns Hopkins University, US
2017 - 2022 - Postdoctoral fellow with Prof. Sarah Woodson, Dept. of Biochemistry, Johns Hopkins University, USA
HONORS, PRIZES, AND AWARDS
2024-2029 EMBO Installation Grant
2023-2028 Sonata Bis grant (National Science Centre)
2023-present Member of the NAR Early Career Researcher Advisory Board
2022 - Invited panelist "Diverse Voices from Rising Scientists", RNA Society meeting, Boulder, USA
2022 - RNA Society Research Presentation Fellowship
2021 - Invited interview with Molecular Cell "Meet the authors", doi: 10.1016/j.molcel.2021.04.011
2021 - Early-career reviewer in Elife (Structural Biology and Molecular Biophysics)
2021 - Conference Award, RNA Society
2019 - Travel Award, RNA Society
2018 - Travelling Fellowship, The Company of Biologists
2015 - present - Member, RNA Society
2015-2018 Preludium grant (National Science Centre)
PUBLICATIONS BY DR. EWELINA MAŁECKA PRIOR TO JOINING IIMCB:
-
Małecka EM, Woodson SA.
RNA compaction and iterative scanning for small RNA targets by the Hfq chaperone.
Nat Commun. 2024 Mar 7;15(1):2069.
doi:10.1038/s41467-024-46316-6.
- Sarni SH, Roca J, Du C, Jia M, Li H, Damjanovic A, Małecka EM, Wysocki VH, Woodson SA.
Intrinsically disordered interaction network in an RNA chaperone revealed by native mass spectrometry.
Proc Natl Acad Sci U S A. 2022 Nov 22;119(47):e2208780119.
doi: 10.1073/pnas.2208780119.
-
Małecka EM, Hua B, Woodson SA.
Single-Molecule FRET Studies of RNA Structural Rearrangements and RNA-RNA Interactions.
Methods Mol Biol. 2022;2518:271-289.
doi: 10.1007/978-1-0716-2421-0_16.
-
Małecka EM, Sobańska D, Olejniczak M.
Bacterial Chaperone Protein Hfq Facilitates the Annealing of Sponge RNAs to Small Regulatory RNAs.
J Mol Biol. 2021 Nov 19;433(23):167291.
doi: 10.1016/j.jmb.2021.167291.
-
Malecka EM, Bassani F, Dendooven T, Sonnleitner E, Rozner M, Albanese TG, Resch A, Luisi B, Woodson S, Bläsi U.
Stabilization of Hfq-mediated translational repression by the co-repressor Crc in Pseudomonas aeruginosa.
Nucleic Acids Res. 2021 Jul 9;49(12):7075-7087.
doi: 10.1093/nar/gkab510.
-
Małecka EM, Woodson SA.
Stepwise sRNA targeting of structured bacterial mRNAs leads to abortive annealing.
Mol Cell. 2021 May 6;81(9):1988-1999.e4.
doi: 10.1016/j.molcel.2021.02.019.
-
Panja S, Małecka EM, Santiago-Frangos A, Woodson SA.
Quantitative Analysis of RNA Chaperone Activity by Native Gel Electrophoresis and Fluorescence Spectroscopy.
Methods Mol Biol. 2020;2106:19-39.
doi: 10.1007/978-1-0716-0231-7_2.
-
Małecka EM, Woodson SA.
Ribosomes clear the way for siRNA targeting.
Nat Struct Mol Biol. 2020 Sep;27(9):775-777.
doi: 10.1038/s41594-020-0495-4.
-
Santiago-Frangos A, Fröhlich KS, Jeliazkov JR, Małecka EM, Marino G, Gray JJ, Luisi BF, Woodson SA, Hardwick SW.
Caulobacter crescentus Hfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc Natl Acad Sci U S A. 2019 May 28;116(22):10978-10987.
doi: 10.1073/pnas.1814428116.
-
Małecka EM, Stróżecka J, Sobańska D, Olejniczak M.
Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq.
Biochemistry. 2015 Feb 10;54(5):1157-70.
doi: 10.1021/bi500741d.
-
Sobkowiak L, Bielewicz D, Malecka EM, Jakobsen I, Albrechtsen M, Szweykowska-Kulinska Z, Pacak A.
The Role of the P1BS Element Containing Promoter-Driven Genes in Pi Transport and Homeostasis in Plants.
Front Plant Sci. 2012 Mar 30;3:58.
doi: 10.3389/fpls.2012.00058.

Group Leader:
Ewelina Małecka, PhD
Postdoctoral Researcher:
Maciej Dylewski, PhD
PhD Students:
Ewa Izdebska
Aiswarya Mohan
Junior Research Scientist:
Daria Demina
Intern:
Sebastian Machera
Msc student:
Zuzanna Grzegorczyk
Laboratory Support Specialist:
Karolina Komorowska
Technician:
Katarzyna Kaca




