We explore how RNA-binding proteins control gene activity and help fight off RNA viruses. Combining techniques from structural biology to live cell experiments, we have discovered RBPs and small molecules that influence production of key protein responsible for Parkinson’s disease. We also identified a new RBP that plays a key role in the immune system’s response to viral infections. Our research not only answers core questions in molecular biology but also opens new paths for treating both infectious and non-infectious diseases.
Research Summary
RNA is a fundamental molecule essential for life, carrying genetic instructions from DNA to build proteins and performing crucial regulatory and catalytic roles. One leading hypothesis suggests that life originated in an RNA world prior to the appearance of DNA. However, RNA depends on RNA-binding proteins (RBPs), which ensure it is properly processed, transported, and translated, supporting genetic information flow and maintaining cell function. RNA-protein interactions also underpin immune defenses; certain RBPs detect virus-derived RNAs, triggering immune responses. Disruption of these interactions contributes to disease, including viral immune evasion and neurodegenerative disorders such as Parkinson’s disease (PD).
At the Dioscuri Centre for RNA-Protein Interactions in Human Health and Disease at the International Institute of Molecular and Cell Biology in Warsaw, we study how these interactions influence cellular systems. Our research addresses two main areas: 1) how RBPs enable immune detection of viral and therapeutic RNAs and 2) how targeting RNA-protein interactions may help treat human diseases, especially viral infections and PD.
We investigate how RBPs recognize features of viral RNA and initiate immune responses by triggering molecules such as interferons. Understanding these processes could support antiviral drug or vaccine development. In PD, we focus on regulation of alpha-Synuclein by RBPs and microRNAs. We study how RNA-protein interactions can be modulated to restore healthy regulation, aiming to influence disease progression at the molecular level.
Scientific Impact
- Advancement in understanding of RNA-protein interactions as key regulators of innate immunity, enabling the development of novel antiviral strategies.
- Identification of molecular mechanisms underlying Parkinson’s disease, revealing novel therapeutic targets.
Future Goals
We aim to employ a multidisciplinary approach to explore the links between RNA biology and human diseases. We will be probing the involvement of RBPs in viral signaling pathways, which can lead to the identification of a novel and broad range of antiviral therapies. Furthermore, we will be expanding our understanding of RNA regulatory pathways and seeking compounds to decrease alpha-Synuclein expression in Parkinson’s disease.
Collaborations
Together with Prof. Juri Rappsilber (Berlin Technical University), we use mass spectrometry for whole proteome studies as well as structural analyses. Prof. Rappsilber is a German partner of the Dioscuri Centre for RNA-Protein Interaction in Human Health and Disease.
Together with Prof. Andrzej Dziembowski (IIMCB) and Prof. Gunther Hartmann (Bonn Medical University), we are investigating the immunogenicity of therapeutic RNAs.
Together with Elżbieta Nowak, PhD, DSc Habil, and Prof. Marcin Nowotny (IIMCB), we are elucidating the structures of RNA-binding proteins (RBPs).
Together with Dr. Katarzyna Mleczko-Sanecka (IIMCB), Wojciech Pokrzywa, PhD, DSc Habil (IIMCB), and Prof. Tilo Kunath (University of Edinburgh), we are elucidating the effects of RBP-targeted compounds in cells and whole organisms.
Comment
"In my research, I unravel the intricate connections between RNA biology and human diseases, aiming to discover innovative approaches for next-generation treatments", says Prof. Gracjan Michlewski
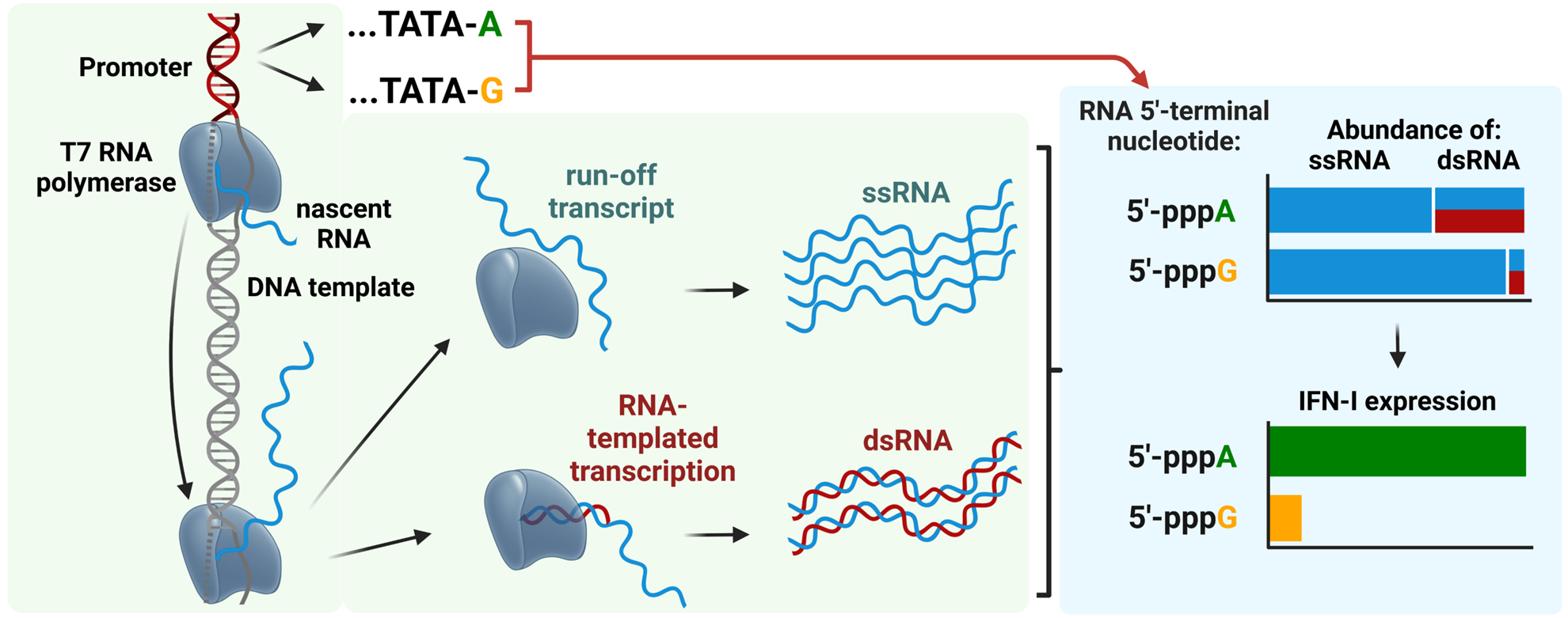
RNAs produced by in vitro transcription with 5′-pppA terminal nucleotide are more immunogenic than those with 5′-pppG due to higher levels of double-stranded RNA (dsRNA) that strongly activate the RIG-I/Interferon type 1 pathway https://doi.org/10.1093/nar/gkae1252.