We explore the universe of RNA, not just as a messenger of genetic blueprints but as a dynamic entity that shapes life at the molecular level. Our work treats RNA as a puzzle, where each piece is a clue to its myriad roles from catalysts to regulators within the cell. With advanced computational models and experimental techniques, we decode RNA's complex structures and interactions, especially with proteins and small molecules.
Research Summary
Our work is focused on the development and applications of new methods for RNA structure determination and modeling RNA interactions, combining computational predictions with experimental analyses. This approach is vital for delving into RNA's role in biological processes, contributing significantly to the fields of molecular biology, bioinformatics, and structural biology. Our contributions include software tools like ModeRNA and SimRNA, which have become essential for researchers globally. We investigate the three-dimensional structures of RNA from viruses, bacteria, and humans, focusing on potential targets for small molecules. Our research is enriched by interdisciplinary collaborations with various research groups and with commercial partners in Poland and worldwide.
Scientific Impact
- Tools for RNA modeling & small molecule interactions
- Determined structures of viral & bacterial RNAs (e.g., coronavirus 5' regions)
- Collaborated with Molecure to validate tools in drug discovery (inhibiting cancer protein translation)
Future Goals
We aim to deepen our understanding of RNA structure and function, particularly by studying molecules with therapeutic potential. Our advances in computational methods, integrated with experimental studies, are geared towards contributing to both fundamental research and future practical applications, with the ultimate goal of positively impacting scientific knowledge and human health.
Collaborations
- Cryo-EM & RNA structure: S. Glatt (Poland), Z. Su (China)
- RNA dynamics & AFM: F. Moreno-Herrero (Spain)
- RNA CD and FTIR: V. Arluison, F. Wien (France)
- RNA-targeting drugs: Molecure (Poland)
Comment
“Our aim is to determine RNA structures and interactions, and design new molecules with functions with potential for applications in medicine and biotechnology”, says Prof. Janusz Bujnicki.
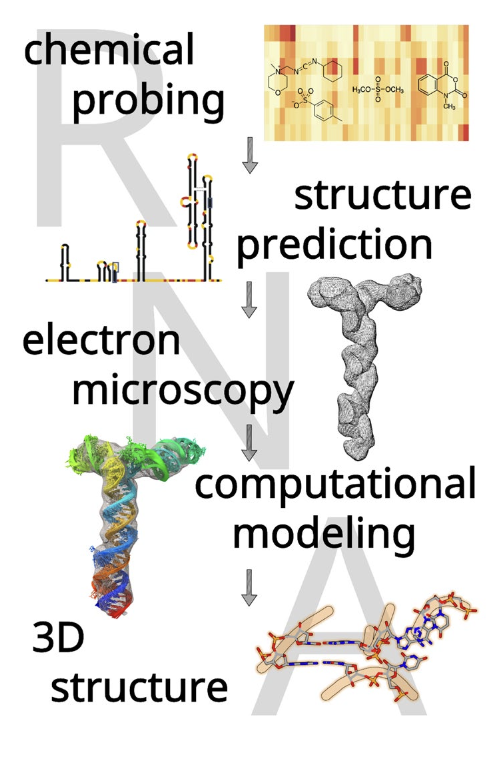
Determination of RNA 3D structure and dynamics, using a combination of computational and experimental methods. Example for the 5′-proximal region in SARS-CoV-2 RNA.
Visit the laboratory website for more details: https://genesilico.pl/
 |
Janusz M. Bujnicki, PhD, Professor
Correspondence address:
Laboratory of Bioinformatics and Protein Engineering
International Institute of Molecular and Cell Biology
4 Ks. Trojdena Street, 02-109 Warsaw, Poland
Email: This email address is being protected from spambots. You need JavaScript enabled to view it.
www: iimcb.genesilico.pl
tel: +48 (22) 597 0750; fax: +48 (22) 597 0715
|
DEGREES
2009 - Professor of Biological Sciences, nomination by the President of the Republic of Poland
2005 - DSc Habil in Biochemistry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland
2001- PhD in Biology, University of Warsaw, Faculty of Biology, Poland
1998 - MSc in Microbiology, University of Warsaw, Faculty of Biology, Poland
PROFESSIONAL EXPERIENCE
2002-present - Professor, Head of Laboratory of Bioinformatics and Protein Engineering, International Institute of Molecular and Cell Biology in Warsaw, Poland (100% appointment)
2019-present - Scientific Advisor, Łukasiewicz Research Network - PORT Polish Center for Technology Development (25% appointment)
2006-2020 - Associate Professor (extraordinarius), Bioinformatics Laboratory, Institute of Molecular Biology and Biotechnology, Adam Mickiewicz University, Poznań, Poland
2010-2011 - Deputy Director, International Institute of Molecular and Cell Biology in Warsaw (1 year rolling position)
2008 - Visiting Professor, University of Tokyo, Japan(sabbatical)
2004-2006 - Assistant Professor, Adam Mickiewicz University, Poznań, Poland
2001 - Visiting Scientist, National Center for Biotechnology Information,National Institutes of Health, Bethesda, Maryland, USA
1999-2002 - Research Scientist, Bioinformatics Laboratory, International Institute of Molecular and Cell Biology in Warsaw, Poland
1998-2000 - Senior Research Assistant, Henry Ford Hospital, Detroit, Michigan, USA
SELECTED PROFESSIONAL AFFILIATIONS
2019-2022 - Member, Committee for Science Evaluation, Ministry of Science and Higher Education
2020 - Member, Advisory Group on Preventing, Counteracting and Combating COVID-19, Ministry of Science and Higher Education
2019-present - Member, University Council, University of Warsaw (Chairman, 2019-2020)
2018-present - Member, Academia Europaea
2018-present - Member, European Molecular Biology Organization
2017-present - Member, European Science Advisors Forum
2016-present - Corresponding Member, Polish Academy of Sciences
2016-2017 - Member, Council of the National Science Congress
2015-2020 - Member, Group of Chief Scientific Advisors, European Commission’s Scientific Advice Mechanism
2014-2018 - Member, Scientific Policy Committee, Polish Ministry of Science and Higher Education
2013-present - Executive Editor, Nucleic Acids Research
2013-2016 - Member, Scientific Committee of the Innovative Medicines Initiative
2013-2015 - Member, Science Europe: Life, Environmental and Geo Sciences (LEGS) Scientific Committee
2011-2016 - Member, Polish Young Academy, Polish Academy of Sciences
2007-present - Member, Polish Bioinformatics Society (founding member; Vice-President, 2007-2010; President, 2011-2013)
2007-present - Member, RNA Society
2001-present - Member, International Society for Computational Biology (Senior Member, 2015-)
SELECTED AWARDS AND FELLOWSHIPS
2019 - André Mischke Young Academy of Europe Prize for Science and Policy Honorary Award "For Merits for Inventiveness," Prime Minister at the request of the Polish Patent Office
2019 - Award for Organizational Achievements, Ministry of Science and Higher Education
2017 - Crystal Brussels Sprout Award
2015 - Jan Karol Parnas Award of the Polish Biochemical Society
2014 - National Science Centre Award for outstanding scientific achievements
2014 -Master Award, Foundation for Polish Science
2014 - Prime Minister’s Award for outstanding scientific achievements
2014 - Selected as one of “25 leaders for the next 25 years” by Teraz Polska magazine of the Polish Promotional Emblem Foundation
2014 - Knight's Cross of the Order of Polonia Restituta
2014 - Award in the Science category of the national plebiscite “Poles with Verve”
2013 - ERC Proof of Concept Grant
2012 - Award for Outstanding Research Achievements, Ministry of Science and Higher Education
2010 - ERC Starting Grant (2011-2015)
2009 - Scholarship for Outstanding Young Scientists, Minister of Science and Higher Education
2009 - Award for Research Achievements, Ministry of Science and Higher Education
2006 - Prime Minister Award for habilitation thesis
2006 - Young Researcher Award in Structural and Evolutionary Biology, Visegrad Group Academies of Sciences
2003, 2004 - START Scholarship for Young Scientists, Foundation for Polish Science
2002-2005 - EMBO/HHMI Young Investigator Award
2002 - Award for best Polish genetics-related publication in 2002, Polish Genetics Society
2001 - Award for best Polish publication on nucleic acid biochemistry in 2000, Polish Biochemical Society and Sigma-Aldrich
DOCTORATES DEFENDED UNDER LAB LEADER’S SUPERVISION
A. Żylicz-Stachula, A. Chmiel, I. Cymerman, A. Czerwoniec, M. Gajda, M. Pawłowski, J. Sasin-Kurowska, J. Kosiński, A. Obarska-Kosińska, S. Pawlak, E. Purta, K. Tkaczuk, Ł. Kościński, M.Rother, W. Potrzebowski, I. Korneta, T. Puton, J. Kasprzak, I. Tuszyńska, Ł. Kozłowski, M. Werner, A. Kamaszewska, A. Philips, K. Milanowska, M. Piętal, D. Matelska, K. Majorek, M. Domagalski, T. Osiński, M. Machnicka, M. Magnus, K. Szczepaniak, M. Zielińska, Astha, I. Foik, D. Toczydłowska-Socha.

Lab Leader:
Senior Researchers:
-
Elżbieta Purta, PhD
-
Filip Stefaniak, PhD
Postdoctoral Researchers:
Research Assistants:
-
Agata Bernat, MSc
-
Dominik Sordyl, MSc
Research Technicians:
-
Masoud Amiri Farsani, MSc
-
Seyed Naeim Moafinejad, MSc
PhD Students:
Lab Technician:
Laboratory Support Specialist:



